Rep:Mod:pg1910z
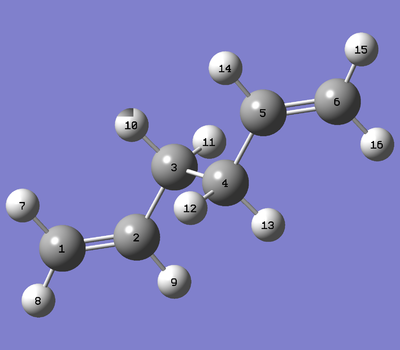
1,5-Hexadiene
Anti-optimisation
| File Name | 15_hexadiene_OP |
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RHF |
| Basis Set | 3-21G |
| Charge | 0 |
| Spin | Singlet |
| E(RHF) | -231.69253529 a.u. |
| RMS Gradient Norm | 0.00001141 a.u. |
| Imaginary Freq | |
| Dipole Moment | 0.0001 Debye |
| Point Group | C1 |
| Job cpu time: | 0 days 0 hours 2 minutes 32.2 seconds. |
| File: | File:15 hexadiene OP.log |
Item Value Threshold Converged?
Maximum Force 0.000017 0.000450 YES
RMS Force 0.000006 0.000300 YES
Maximum Displacement 0.000747 0.001800 YES
RMS Displacement 0.000203 0.001200 YES
Predicted change in Energy=-1.360587D-08
Optimization completed.
-- Stationary point found.
| |
When symmeterised point group goes to Ci
Low frequencies --- -2.3512 -1.2713 -0.0007 -0.0006 -0.0002 1.1743 Low frequencies --- 71.2984 85.7194 116.2266
Gauche

| File Name | 15_hexadiene_gauche3_OP |
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RHF |
| Basis Set | 3-21G |
| Charge | 0 |
| Spin | Singlet |
| E(RHF) | -231.69266115 a.u. |
| RMS Gradient Norm | 0.00001617 a.u. |
| Imaginary Freq | |
| Dipole Moment | 0.3399 Debye |
| Point Group | C1 |
| Job cpu time: | 0 days 0 hours 1 minutes 49.9 seconds. |
| File | File:15 hexadiene gauche OP.log |
Item Value Threshold Converged?
Maximum Force 0.000051 0.000450 YES
RMS Force 0.000013 0.000300 YES
Maximum Displacement 0.001749 0.001800 YES
RMS Displacement 0.000637 0.001200 YES
Predicted change in Energy=-7.279818D-08
Optimization completed.
-- Stationary point found.
| |
Low frequencies --- -1.5569 -0.5609 -0.0009 -0.0007 -0.0007 1.3988 Low frequencies --- 74.6163 105.0624 130.5818
1,5-Hexadiene Gauche symmetric

| File Name | 15_hexadiene_gauche_OP |
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RHF |
| Basis Set | 3-21G |
| Charge | 0 |
| Spin | Singlet |
| E(RHF) | -231.69153036 a.u. |
| RMS Gradient Norm | 0.00000457 a.u. |
| Imaginary Freq | |
| Dipole Moment | 0.1281 Debye |
| Point Group | C1 |
| Job cpu time: | 0 days 0 hours 4 minutes 8.9 seconds. |
| File | File:15 hexadiene gauche2 OP.log |
Item Value Threshold Converged?
Maximum Force 0.000012 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.000464 0.001800 YES
RMS Displacement 0.000110 0.001200 YES
Predicted change in Energy=-1.625564D-09
Optimization completed.
-- Stationary point found.
| |
When symmeterise operation preformend the point group becomes C2
Low frequencies --- -1.5338 -0.0010 -0.0007 0.0003 0.5021 1.3597 Low frequencies --- 65.5170 68.6456 147.3426
File:PG 15 hexadiene gauche2 FR.log
Matches up with Gauch 4 structure
Gauche 1 arrangement

| File Name | 15_hexadiene_gauche1_O |
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RHF |
| Basis Set | 3-21G |
| Charge | 0 |
| Spin | Singlet |
| E(RHF) | -231.68771613 a.u. |
| RMS Gradient Norm | 0.00002137 a.u. |
| Imaginary Freq | |
| Dipole Moment | 0.4556 Debye |
| Point Group | C1 |
| Job cpu time: | 0 days 0 hours 3 minutes 42.5 seconds. |
| File: | File:PG 15 hexadiene gauche1 OP.log |
Item Value Threshold Converged?
Maximum Force 0.000046 0.000450 YES
RMS Force 0.000012 0.000300 YES
Maximum Displacement 0.001463 0.001800 YES
RMS Displacement 0.000428 0.001200 YES
Predicted change in Energy=-3.135658D-08
Optimization completed.
-- Stationary point found.
| |
Low frequencies --- -3.0201 -2.2455 -0.6366 -0.0010 -0.0009 -0.0009 Low frequencies --- 113.9867 137.4299 152.3309
File:PG 15 hexadiene gauche1 FR.log
When symmeterise operation preformed the point group becomes C2
Comparison of Structures
| Structure | Point Group | Calculated Energy | Relative Energy |
|---|---|---|---|
| Anti2 | Ci | -231.69253529 | |
| Gauche3 | C1 | -231.69266115 | 0.00 |
| Gauche4 | C2 | -231.69153036 | |
| Gauche1 | C2 | -231.68771613 |
Anti-optimisation

| 15_hexadiene_OPDFT3 | |
| File Name | 15_hexadiene_OPDFT3 |
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d) |
| Charge | 0 |
| Spin | Singlet |
| E(RB3LYP) | -234.61171047 a.u. |
| RMS Gradient Norm | 0.00006317 a.u. |
| Imaginary Freq | |
| Dipole Moment | 0.0000 Debye |
| Point Group | CI |
| Job cpu time: | 0 days 0 hours 1 minutes 21.1 seconds. |
| File: | File:PG 15 hexadiene OPDFT3.log |
Item Value Threshold Converged?
Maximum Force 0.000138 0.000450 YES
RMS Force 0.000030 0.000300 YES
Maximum Displacement 0.001096 0.001800 YES
RMS Displacement 0.000418 0.001200 YES
Predicted change in Energy=-2.058476D-07
Optimization completed.
-- Stationary point found.
| |
Low frequencies --- -9.9447 -0.0009 -0.0007 -0.0005 3.2271 10.6991 Low frequencies --- 73.9125 80.8531 121.3516
File:PG 15 hexadiene OPDFT3 FR.log
On first glances, the two structure appear the same, however on closer looking analysis there does appear to be some differences in bond lengths, angles and dihedral angles, which while on the surface seem to be minor, do show the effect that using different calculation methods and basis sets can make a difference to more then just the energy calculations.
| Parameter | HF/3-21G | DFT/6-31G* |
|---|---|---|
| 1-2 length (Å) | 1.316 | 1.333 |
| 1-7 length (Å) | 1.073 | 1.089 |
| 2-9 length (Å) | 1.077 | 1.092 |
| 1,2,3 angle ( °) | 124.8 | 125.3 |
| 2,3,4 angle ( °) | 111.3 | 112.7 |
| 7,1,2,3 angle ( °) | 1.1 | 0.7 |
| 9,2,3,10 angle ( °) | 174.3 | 177.0 |
While the accuracy of the calculations mean that the difference between some of the parameters on the table are in actuality not truly valid comparisons. I felt it useful to still note their values to show a difference. The main parameter that I do feel worth pointing out is the last one, the dihedral angle between hydrogens 9 and 10, as well as the slight twisting of the double bone. I feel unable to rationalise the reason that the structural parameters as they relate to the structure itself (i.e. why the double bond has a twist, and why the the anti-periplanar hydrogens deviate from 180°.) however the difference between the two calculation is due to the different methods used to run the calculations which will change how the the orbitals are modeled as well as the basis sets, which will change which orbitals are being calculated rather then using assumed densities, and to what detail the calculations are run at.
ThermoChemistry
Temperature 298.150 Kelvin. Pressure 1.00000 Atm. ...
Sum of electronic and zero-point Energies= -234.469202 Sum of electronic and thermal Energies= -234.461855 Sum of electronic and thermal Enthalpies= -234.460910 Sum of electronic and thermal Free Energies= -234.500782
Gaussian was unable to cope with the input of Temperature = 0K (ignoring the command entirely) so instead 0.0001K was inputted and the expected result of a loss of thermal energies, enthalpies and free energy was seen.
Temperature 0.000 Kelvin. Pressure 1.00000 Atm. ...
Sum of electronic and zero-point Energies= -234.469202 Sum of electronic and thermal Energies= -234.469202 Sum of electronic and thermal Enthalpies= -234.469202 Sum of electronic and thermal Free Energies= -234.469202
File:PG 15 hexadiene OPDFT3 FR 0.0001K.log
Cope-Rearrangement
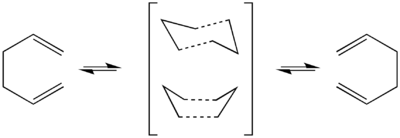
The cope rearrangement, at its most basic level is a [3,3]-sigmatropic rearrangement of a 1,5-diene. It is well accepted that the rearrangement can take place, either via a boat or chair transition state (TS). It is impossible by experimentation or observation to isolate or analyses the TS, however using computational techniques it is possible to model the reaction, and compute a probable TS and by arranging the fragments it is possble to force the reaction to go via one or the other. We shall try several different methods to get to the TS. Each method has its benefits as well as its pitfalls. For the most part, the pitfalls aren't noticeable in these calculations due to the level of calculation and the number of atoms in the calculation. First we shall look at the chair TS.
Chair Transition State
The first method is done using computed force constants. The main Thing here being to arrange the fragment into what is thought to be the TS of the reaction.

Step 1: Optimise the structure of one of the allyl (CH2-CH-CH2) fragments at HF/3-21G level of theory. It is not necessary to use a higher level of theory yet, as the aim at the moment is to see if there is indeed a TS seen. If necessary once we find a TS these calculations can be re-done at a higher level of theory.
Step 2: Arrange two allyl fragment one atop the other, with the center carbons facing opposite directions. The end carbons of one fragment should sit over the end carbons of the other both with a gap of around 2.2Å. This has been done in the picture shown, and when viewed correctly, it is possible to make out the chair like arrangement of carbons.
Step 3: Carry out a freq+opt to TS(Berny) using the same HF/3-21G level of theory. This calculates the second derivative of the energy curvature as the molecule is being optimised, ensuring that the energies along the forming bond are increasing in energy to a maximum, the force constant only needs to be calculated once and the keyword opt=NoEigan is added to allow the computer to cope with the possibility of more then one imaginary frequency. The resulting molecule should hopefully be a rough representation of the final product.
This next step involves taking the atoms that will be forming the bond and freezing their coordinates in space, the rest of the molecule is then optimised to a minimum. Causing all of the surrounding molecule to assume a relaxed structure. After this, the bond forming atoms are unfrozen, and then used as the coordinates along which the computation is looking for the energy to increase to a maximum.
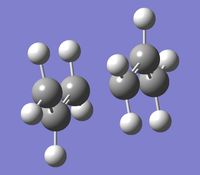
Step 1: Using the redundant coordinate editor the carbon atoms that, between which a bond will form are selected in their bond forming pairs, and the freeze coordinate option is selected. The molecule is then Optimised to a Minimum using HF/3-21G adding the keyword Opt=ModRedundant. The effect of this step is seen, where before the fragments were both planer, what is now seen is the hydrogen attached to either end of the carbons spread away from each other, assuming a more SP3 like structure.
Step 2: From the log file from step 1, Unfreeze the coordinates and set it to derivative, submitting the job optimising to the transition state, with no force constant calculations. Out of this we expect to find the TS similar to earlier.
Bellow is a comparison of the two approaches.
| heading | heading |
|---|---|
| cell | cell |
| cell | cell |
| Property | Method 1 | Method 2 |
|---|---|---|
| Guesses | 15 | 10 + 6 |
| E(RHF) (a.u.) | -231.61932239 | -231.61932232 |
| imaginary freq (Hz) | -817.92 | -817.89 |
| RMS(gradient) | 0.00002313 | 0.00002217 |
| Bond Length (Å) | 2.020 | 2.021 |
There seems to be no noticeable difference between the two methods, at lest at the size of system and theory level that we are looking.
Boat Transition state
The boat transition state is analysed using yest another optimisation method called QTS2. This works by giving Gaussian the molecule in the start configuration and again in its final configuration. What the calculation then attempts to do is to figure out the TSof this particular transformation.
 |
|
The set up for the calculation is quite simple. The Anti2 optimised hexadiene structure is copied into a new file and the geometry adjusted so that it is in a boat like arrangement with the two ene- groups in bond forming proximity (~2.2 Å). This structure is then copied and added as a new molecule group, to aid the labeling process, they can be arranged as shown so that they mirror one another. With the aid of the Atom list console the atoms are relabeled similar to the ones shown to the right, ensuring that all the atoms in relative space retain their label from reactant to product.

The calculation is set up as a QTS2', Freq+Opt using HF/3-21Gtheory level.
| File Name | PG_Trans_BOAT_2_OPFR |
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RHF |
| Basis Set | 3-21G |
| Charge | 0 |
| Spin | Singlet |
| E(RHF) | -231.60280219 a.u. |
| RMS Gradient Norm | 0.00004199 a.u. |
| Imaginary Freq | 1 |
| Dipole Moment | 0.1582 Debye |
| Point Group | C1 |
| Job cpu time: | 0 days 0 hours 1 minutes 3.7 seconds. |
| File: | File:PG BoatTS OPFR.log |
Item Value Threshold Converged?
Maximum Force 0.000079 0.000450 YES
RMS Force 0.000020 0.000300 YES
Maximum Displacement 0.001208 0.001800 YES
RMS Displacement 0.000368 0.001200 YES
Predicted change in Energy=-3.056240D-07
Optimization completed.
-- Stationary point found.
| |
Low frequencies --- -839.6295 0.0005 0.0006 0.0007 4.5588 5.9315 Low frequencies --- 6.7417 155.4945 381.5944 | |
The output is very similar to those from the previous set of TS calculations, and the boat like structure can clearly be determined.
IRC
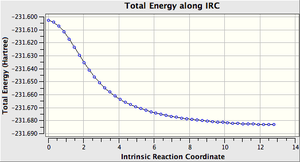
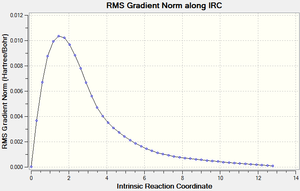
While the molecule that the inputted file was based on was the anti2 structure, that does not necessarily mean that it is the structure most able to go through the calculated TS especially considering the amount of modification performed to get it near to the boat like structure. what we can do is use the IRC job which will attempt to follow the the steepest gradient of energy when altering the geometry of the molecule in small steps, in effect finding the products/reactants for the calculated TS

| File Name | PG_Trans_BOAT_2_OPFR_Tryagain_chk_IRC |
| File Type | .log |
| Calculation Type | IRC |
| Calculation Method | (RHF) |
| Basis Set | (3-21G) |
| Charge | 0 |
| Spin | Singlet |
| E(RHF) | -231.60280219 a.u. |
| RMS Gradient Norm | 0.00004199 a.u. |
| Imaginary Freq | |
| Dipole Moment | 0.1582 Debye |
| Point Group | C1 |
| Job cpu time: | 0 days 0 hours 12 minutes 53.1 seconds. |
| File: | File:PG BoatTS IRC.log |
PES minimum detected on this side of the pathway.
Magnitude of the gradient = 0.0001496
Calculation of FORWARD path complete.
Reaction path calculation complete.
Energies reported relative to the TS energy of -231.602802
-------------------------------------------------------------------
Summary of reaction path following
-------------------------------------------------------------------
Energy RxCoord
1 0.00000 0.00000
...
45 -0.08018 12.78782
-------------------------------------------------------------------
Total number of points: 44
Total number of gradient calculations: 45
| |
What we find is that for the Boat TS the structure that will undergo this transformation is the Gauche 3 structure.
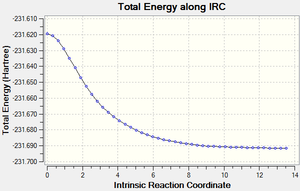
The same procedure is carried out for the Chair TS
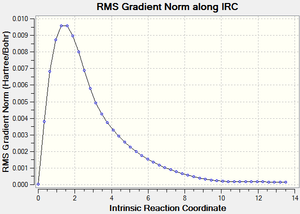
| File Name | PG_Trans_Chair_IRC |
| File Type | .log |
| Calculation Type | IRC |
| Calculation Method | (RHF) |
| Basis Set | (3-21G) |
| Charge | 0 |
| Spin | Singlet |
| E(RHF) | -231.61932239 a.u. |
| RMS Gradient Norm | 0.00002313 a.u. |
| Imaginary Freq | |
| Dipole Moment | 0.0004 Debye |
| Point Group | C1 |
| Job cpu time: | 0 days 0 hours 24 minutes 58.7 seconds. |
| File: | File:PG ChairTS IRC.log |
PES minimum detected on this side of the pathway.
Magnitude of the gradient = 0.0001429
Calculation of FORWARD path complete.
Reaction path calculation complete.
Energies reported relative to the TS energy of -231.619322
-------------------------------------------------------------------
Summary of reaction path following
-------------------------------------------------------------------
Energy RxCoord
1 0.00000 0.00000
...
44 -0.07226 13.51630
-------------------------------------------------------------------
Total number of points: 43
Total number of gradient calculations: 44
| |
We can see from the IRC output, that the structure most likely to go through this TS is the Gauche 2 structure
Activation energies
Now we have found the Boat TS and the Chair TS and we know which structures connect to those TSs we can compare relative energies to find the activation energy of both transformations and the difference in energy between the two transformations. First however to get more accurate values for energy, all the calculations will be recalculated using DFT/6-31G* level of theory. This should hopefully be detailed enough to get improved calculations without introducing unnecessary levels of complications this will only server to slow the calculations down.
| Boat | Chair | |
|---|---|---|
| File Name | PG_BoatTS_2_OPFR_631Gd | PG_Trans_Chair_OP_631G_2 |
| File Type | .log | .log |
| Calculation Type | FREQ | FREQ |
| Calculation Method | RB3LYP | RB3LYP |
| Basis Set | 6-31G(d) | 6-31G(d) |
| Charge | 0 | 0 |
| Spin | Singlet | Singlet |
| E(RB3LYP) | -234.54309307 a.u. | -234.55693218 |
| RMS Gradient Norm | 0.00000094 a.u. | 0.00000062 |
| Imaginary Freq | Singlet | Singlet |
| Dipole Moment | 0.0613 Debye | 0.0000 |
| Point Group | C1 | C2 |
| Job cpu time: | 5 minutes 38.8 seconds. | 18 minutes 24.1 seconds. |
| File: | File:PG BoatTS 2 OPFR 631Gd.log | File:PG ChairTS OP 631G 2.log File:PG Trans Chair OP 631Gd FR.log |
| HF/3-21G | B3LYP/6-31G* | |||||
|---|---|---|---|---|---|---|
| Electronic energy | Sum of electronic and zero-point energies | Sum of electronic and thermal energies | Electronic energy | Sum of electronic and zero-point energies | Sum of electronic and thermal energies | |
| at 0 K | at 298.15 K | at 0 K | at 298.15 K | |||
| Boat TS | -231.60280219 | -231.450936 | -231.445306 | -234.54309307 | -234.402342 | -234.396008 |
| Chair TS | -231.61932239 | -231.466706 | -231.461346 | -234.55693218 | -234.414879 | -234.408950 |
| anti-2 | -231.69253529 | -231.539540 | -231.532566 | -234.61171047 | -234.469202 | -234.461855 |
| HF/3-21G | B3LYP/6-31G* | ||||
|---|---|---|---|---|---|
| at 0 K | at 298.15 K | at 0 K | at 298.15 K | Expt 0K | |
| Boat TS | 232.63 | 229.10 | 175.54 | 172.88 | |
| Chair TS | 191.22 | 186.99 | 142.63 | 138.90 | |
1 hartree = 2625.5 kJ/mol (kilojoules per mole)
Diels Alder Cyclo-addition
Here were will attempt to model the TS and the reactions energies of a massively important reaction in the world of chemistry.
Starting Simple
Here we are going to take on molecule of ethene and one of cis-butydiene, and using a semi-emperical method with the AM! basis set, we shall model the molecular orbitals of the two in the hopes of better understanding the HOMO/LUMO orbitals and how they might interact.
First using the same method and basis set, we must optimise the molecule.
| File Name | PG_DiENE_HE_AM1_OP | PG_ENE_OP |
| File Type | .log | .log |
| Calculation Type | FOPT | FOPT |
| Calculation Method | RAM1 | RAM1 |
| Basis Set | ZDO | ZDO |
| Charge | 0 | 0 |
| Spin | Singlet | Singlet |
| E(RAM1) | 0.04879717 a.u. | -78.58745830 a.u. |
| RMS Gradient Norm | 0.00000068 a.u. | 0.00000480 a.u. |
| Imaginary Freq | ||
| Dipole Moment | 0.0415 Debye | 0.0000 Debye |
| Point Group | C2V | C1 |
| Job cpu time: | 0 hours 0 minutes 18.0 seconds. | 0 hours 0 minutes 32.4 seconds. |
| File: | File:PG DiENE HE AM1 OP.log | PG_ENE_OPAM1.log |
Item Value Threshold Converged?
Maximum Force 0.000001 0.000015 YES
RMS Force 0.000000 0.000010 YES
Maximum Displacement 0.000010 0.000060 YES
RMS Displacement 0.000004 0.000040 YES
Predicted change in Energy=-1.294434D-11
Optimization completed.
-- Stationary point found.
|
Item Value Threshold Converged?
Maximum Force 0.000010 0.000015 YES
RMS Force 0.000006 0.000010 YES
Maximum Displacement 0.000026 0.000060 YES
RMS Displacement 0.000016 0.000040 YES
Predicted change in Energy=-6.132474D-10
Optimization completed.
-- Stationary point found.
|
Energy Calculation.
File:PG DiENE HE AM1 EN2.log | File:PG ENE OP ENAM1.log
| Orbital | Ethene | Butadiene |
|---|---|---|
| HOMO | 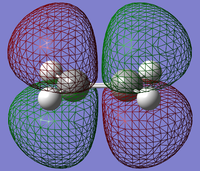 |

|
| LUMO |  |

|
From the diagrams We should expect the LUMO of the Diene to interact with the HOMO of the Ethene (both of which are symmetric in respect to the plane perpendicular to their central bond) , and the HOMO with the LUMO therefor both can be allowed transitions (antisymmetric with respect to plane).
Transition state, Method 1
A guess was made of the Transition state by taking the gaussian template structure of bicyclo-2,2,2-octane, removing one of the -CH2-CH2- units,


| File Name | PG_RXN_T3_AM1_OP_TSb4 |
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RAM1 |
| Basis Set | ZDO |
| Charge | 0 |
| Spin | Singlet |
| E(RAM1) | 0.11165465 |
| RMS Gradient Norm | 0.00000111 |
| Imaginary Freq | 1 |
| Dipole Moment | 0.5605 |
| Point Group | C1 |
| Job cpu time: | 0 days 0 hours 0 minutes 54.1 seconds. |
| File: | File:PG RXN T3 AM1 OP TSb4.log |
Item Value Threshold Converged?
Maximum Force 0.000003 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000144 0.001800 YES
RMS Displacement 0.000038 0.001200 YES
Predicted change in Energy=-4.160235D-10
Optimization completed.
-- Stationary point found.
| |
In the Orbital Diagram of the transition state we see that the interaction seems to be for the LUMO of Ethene and the HOMO of the Diene, this makes sense when remembering that stability increases as conjugation increases, therefor the HOMO of Diene will have a lower energy the the HOMO of Ethene.
Calculating with a higher basis set
Issues arose when trying to run the above using DFT/6-31Gd, as what would often happen is that either only a single bond would form between the two molecules, or the Ethene would completely reorientate to form some bizzar interaction with just one of the double bonds of the Diene-
The Decision was made to run the calculation using QTS2 as it allows us to define the product, therefore forcing the two desired bonds to be formed, in a concerted manner in the transition state.
| File Name | PG_RXN_T2_OP_qts_log_87530 |
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d) |
| Charge | 0 |
| Spin | Singlet |
| E(RB3LYP) | -234.54389656 a.u. |
| RMS Gradient Norm | 0.00000068 a.u. |
| Imaginary Freq | 1 |
| Dipole Moment | 0.3943 Debye |
| Point Group | C1 |
| Job cpu time: | 0 days 0 hours 4 minutes 55.2 seconds. |
| File: | File:PG RXN T2 OP qts.log |
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000032 0.001800 YES
RMS Displacement 0.000008 0.001200 YES
Predicted change in Energy=-3.907341D-11
Optimization completed.
-- Stationary point found.
| |
| Energy File: | File:PG RXN T2 OP qts EN.log File:PG RXN T2 OP qts EN.fchk |
| Frequency at 0K | File:PG RXN T2 OP qts FR0k.log |
| Frequency at 298K | File:PG RXN T2 OP qts FR298k.log |
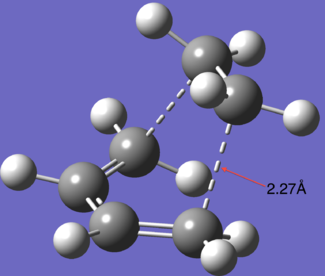
| Parameter | Distance |
|---|---|
| Van der waals radius of Carbon |
1.7 Å |
| C--C Bond length | 1.54 Å |
| C==C Bond Length | 1.34 Å |
| TS C C distance | 2.27 Å |
What we can see with the above information is that the partially formed bond is much to long to be considered a formal bond (comparing it to the C--C and C==C bond lengths) but the fact that the distance is less then twice the van der waals radius of carbon we can conclude that there is some kind of interaction taking place between the fragments.
 |
 |
Here we see that for the Imaginary frequency, that corresponds to the vibration associated to the formation of the bonds in the reaction, both of the bonds are being formed at the same time. This agrees with the accepted idea that the reaction is a pericyclic process.
The lowest positive frequency is just a vibration of the structure and not in itself associated with bond forming, if anything the motion would act to disrupt the bond formation as the twisting motion would break the symmetry of the process.
Cyclohexa-1,3-diene reacting with maleic anhydride
 Seen above is the reaction between Cyclohexadiene and maleic anhydride.
By using computation methods we hope to be able to compare the relitive stabilities between the endo and exo conformers of the reaction product.
Seen above is the reaction between Cyclohexadiene and maleic anhydride.
By using computation methods we hope to be able to compare the relitive stabilities between the endo and exo conformers of the reaction product.
All the calculations have first been run at HF/3-21G and then reoptimised at DTF/6-31G* all properties disscussed, unless otherwise stated, will be from the final optimisation (6-31G*)
Transition states
Due to issues encountered in earlier sections, It was decided to use the QTS2 method to calculate the TS.
| Exo | Endo | |
|---|---|---|
 |

| |
| File Name | PG_ExoRxn_FROP_DFT631GdT3_log_87649 | PG_EndoRxn_FROP_DFT631Gd_log_87564 |
| File Type | .log | .log |
| Calculation Type | FREQ | FREQ |
| Calculation Method | RB3LYP | RB3LYP |
| Basis Set | 6-31G(d) | 6-31G(d) |
| Charge | 0 | 0 |
| Spin | Singlet | Singlet |
| E(RB3LYP) | -612.67931096 a.u. | -612.68339677 a.u. |
| RMS Gradient Norm | 0.00000166 a.u. | 0.00000435 a.u. |
| Imaginary Freq | 1 | 1 |
| Dipole Moment | 5.5502 Debye | 6.1143 Debye |
| Point Group | C1 | C1 |
| Job cpu time: | 0 hours 25 minutes 37.5 seconds. | 0 hours 26 minutes 2.5 seconds. |
| File: | File:PG ExoRxn FROP 631GdT3.log | File:PG EndoRxn FROP 631Gd.log |
By looking at the frontier orbital of the TS we can see that in both the Exo and the Endo TS are an interaction of the HOMO of the cyclohexadiene and the LUMO of the Maleic anhydride. both of these orbitals are antisymetric with respect to both σv so are able to interact.
The orbitals of the (O=C)O(C=O) moiety in the TS seem to be anti-symmetric to the double bond in the maleic anhydride. The two p-orbitals, pointing out of the plane of the molecule are twisted from perpendicular and are out of phase with respect to each other and the oxygen the is part of the five membered ring has it's p-orbital parallel to the double bond but in phase with the two bond forming orbitals. There is a nodal plane in the same place as the mirror place of the molecule
A reordering of the energies of the maleic anhydride orbitals took place when the calculations were done using either HF/3-21G or DFT/6-31Gd, this might be due to the fact that the semi-empirical method doesn't calculate the specific orbital interactions but uses known values and adds them together. When the orbital calculations are run using a higher basis set, the orbital shown in the table becomes the HOMO-2, this maybe just a quirk of the calculation, however it may also be due to the interaction of the double bond with the (O=C)O(C=O) moiety which causes a stablisation of this orbital.
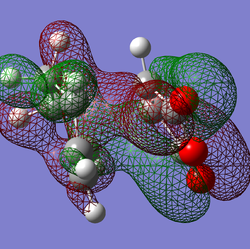
Secondary Orbital Overlap?
 |
 |
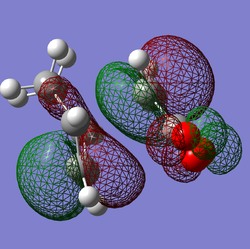 |
Shown above are the representations of orbitals 46, 43 and 37 each of which seem to suggest a potential secondary orbital overlap taking place in the transition state. MO 46 suggests a through space interaction between the carbonyl oxygens and a delocalised orbital encompassing the double bonds of the cyclohexadiene. MO 43 shows again a through space interaction this time from the oxygen in the ring of maleic anhydride. MO 37 shows an orbital that connects the bottom face of maleic anhydride and the top face of the cyclohexadiene, however this orbital is repetitively low in energy from the frontier orbitals.
This being said, the visualized orbitals do not necessarily mean these interactions play any role in the stability of the transition state. After looking through the NBO analysis of the TS there seems to be no significant interactions between the carbonyls (oxygen and carbon) of maleic anhydride and the double bonds of cyclohexadiene. The conclusion being that contraty to the pictographic representations of the orbitals, there is no significant secondary interaction taking place in the 'TS.
Possible strain seen in the endo product
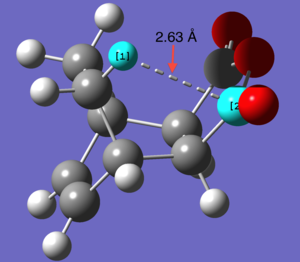
| perameter | Length ( Å) |
|---|---|
| Van der waals radius of Hydrogen |
1.2 |
| Van der waals radius of Carbon |
1.7 |
| Distance between H C | 2.63 |
What we see when looking at the distance between the Hydrogen and Carbon (indicated in the picture to the right) is that distance between the two atoms is less than the sum of their van der waals radii. While the degree to which they interact would be hard to determine as, especially for a carbonyl carbon, there will be effects on the van der waals radius by neighboring atoms.
Reaction Energies
| B3LYP/6-31G* | |||
|---|---|---|---|
| Electronic energy | Sum of electronic and zero-point energies | Sum of electronic and thermal energies | |
| at 0 K | at 298.15 K | ||
| Maleic anhydride | -379.28954470 | -379.233657 | -379.228473 |
| Cyclohexadiene | -233.41893632 | -233.296105 | -233.290933 |
| ExoTS | -612.67931096 | -612.498013 | -612.487662 |
| EndoTS | -612.68339677 | -612.502141 | -612.491787 |
| ExoProd | -612.75578550 | -612.572070 | -612.562604 |
| EndoProd | -612.75829021 | -612.569381 | -612.559977 |
| Activation energy |
Energy change Of Reaction | |||
|---|---|---|---|---|
| 0K | 298.15K | 0K | 298K | |
| Exo | 83.4 | 83.3 | -111.1 | -113.4 |
| Endo | 72.5 | 72.5 | -104.0 | -106.5 |
From the data we see that the Endo form of the product has a smaller activation energy, so will be the kinetic product of the reaction, however when comparing the energy of formation we see that the Exo- product is actually more stable.
IRC
 File:PG EndoRxn FROP DFT631Gd IRC.log |
 File:PG ExoRxn FROP DFT631GdT3 IRC.log |
Above are the resulats from the IRC caculations run on the two TS. In this context, they are not particularly usefull, however they are nice to look at and confirm the reaction path.
Considerations
All these calculations are done as though it is taking place in a vacuum, there for there are no solvent interactions involved. It is known that the Diels-alder reactions runs significantly faster with water then organic solvents alone, therefor this suggests that there might be large solvent effects in this reaction.
It is generally known that the TS of this reaction is aromatic. Given more time, it should be possible to show this to be true by using NICS analysis in Gaussian. By placing ghost atoms in the center of the transition state, you would expect to see a large shielding effect when doing an NMR calculation.