Rep:Mod:nyx2518
NH3 optimisation
Molecule name: NH3
Calculation method: RB3LYP
Basis set: 6-31G(d.p)
Final energy E(RB3LYP) in atomic units(au): -56.55776873 au
What is the RMS gradient(au)? 0.00000485 au
the point group of your molecule: C3v
optimised N-H bond distance: 1.02Å
optimised H-N-H bond angle: 106°
Item table of NH3:
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES
File:NYXPHUNT NH3 OPTF POP.LOG
Jmol dynamic image:
NH3 |
The optimisation file is liked to File:NYXPHUNT NH3 OPTF POP.LOG
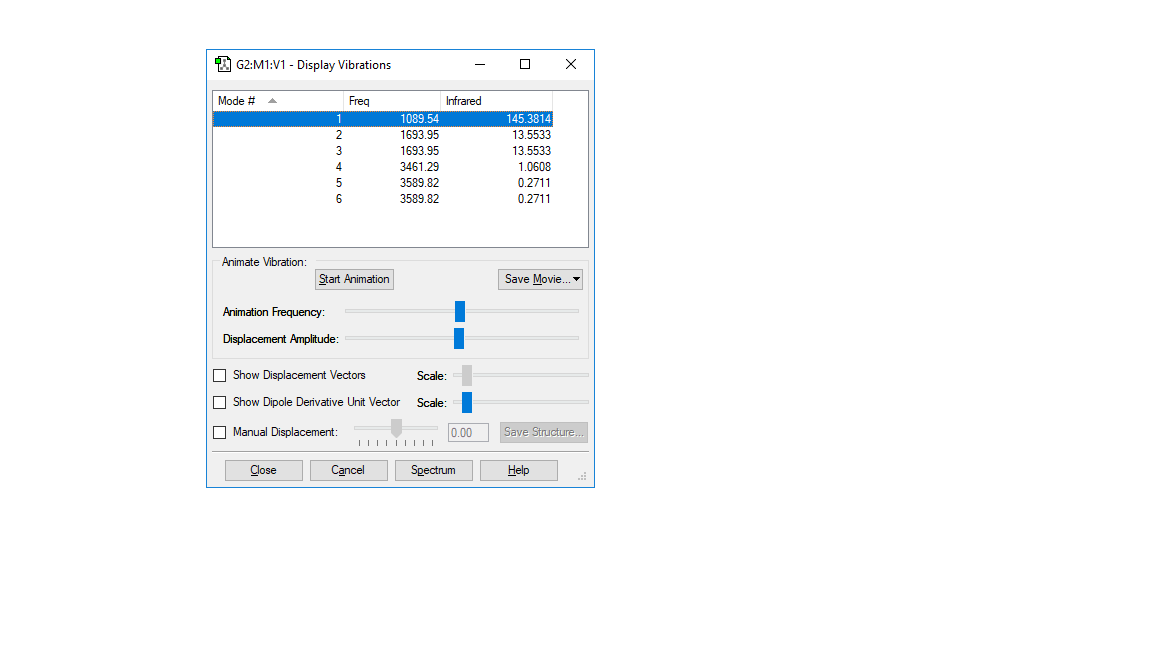
HN3 vibration:
| wavenumber cm-1 | 1090 | 1694 | 1694 | 3461 | 3590 | 3590 |
|---|---|---|---|---|---|---|
| symmetry | A1 | E | E | A1 | E | E |
| intensity arbitrart units | 145 | 14 | 14 | 1 | 0 | 0 |
how many modes do you expect from the 3N-6 rule? 6 modes
which modes are degenerate (ie have the same energy)? 1694 cm-1 and 1694 cm-1, 3590 cm-1 and 3690 cm-1
which modes are "bending" vibrations and which are "bond stretch" vibrations? 1090 cm-1 and 1694 cm-1, 1694cm-1 are bending vibations. 3461 cm-1, 3461 cm-1, 3590 cm-1 and 3590 cm-1 are stretching vibarations.
which mode is highly symmetric? 1090 cm-1 and 3461 cm-1
one mode is known as the "umbrella" mode, which one is this? 1090 cm-1
how many bands would you expect to see in an experimental spectrum of gaseous ammonia? 2
NBO charges:
Charge for N: -1.125, charge for H: +0.375
N2 optimisation
Molecule name: N2
Calculation method: RB3LYP
Basis set: 6-31G(d.p)
Final energy E(RB3LYP) in atomic units(au):-109.52412868 au
What is the RMS gradient(au)? 0.0000006 au
the point group of your molecule: D*H
optimised N-N bond distance: 1.11Å
Item table:
Item Value Threshold Converged? Maximum Force 0.000001 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES
jmol diagram:
N2 |
The optimisation file is liked to File:NYXPHUNT N2 OPTF POP.LOG
N2 vibration:
| wavenumber cm-1 | 2457 |
|---|---|
| symmetry | SGG |
| intensity arbitrart units | 0 |
NBO charges:
Charge for N1: 0, charge for N2: 0
H2 optimisation
Molecule name: H2
Calculation method: RB3LYP
Basis set: 6-31G(d.p)
Final energy E(RB3LYP) in atomic units(au): -1.15928020 au
What is the RMS gradient(au)? 0.09719500 au
the point group of your molecule: D*H
optimised H-H bond distance: 0.74Å
Item table:
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES
jmol diagram:
H2 |
The optimisation file is liked to File:NYXPHUNT H2 OPTF POP.LOG
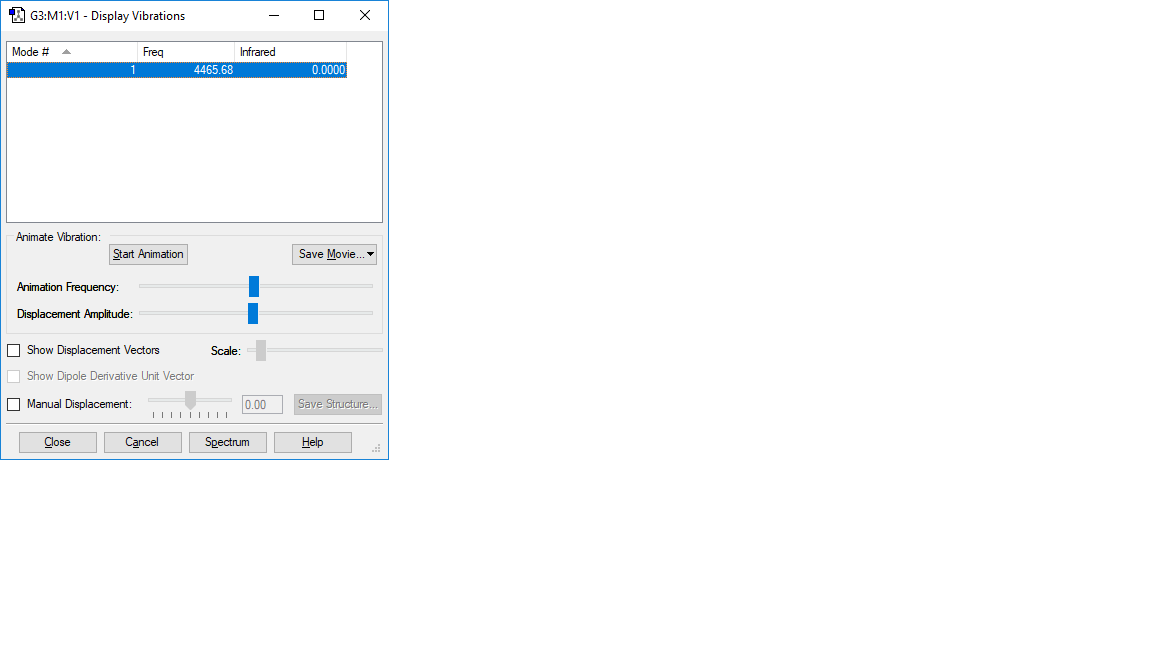
H2 vibration:
| wavenumber cm-1 | 4466 |
|---|---|
| symmetry | SGG |
| intensity arbitrart units | 0 |
NBO charges:
Charge for H1: 0, charge for H2: 0
unique identifier of the mono-metallic TM complex: DAMSUG
The N-N distance obtained from the DAMSUG structure is 1.128Å, which is larger than the computational value (1.11Å). In the crystal stucture, The lone pair on central transition metal are transferred to the empty orbital of N2, which is the LUMO of the N2. Therefore the bond order of nitrogen decrease and the triple bond character of nitrogen decrease (N2 triple bond is weaker than before), the bond distance of N2 is lower than the computational value.
Haber-Bosch reaction energy calculation
E(NH3)= -56.55776873 au
2*E(NH3)= -113.1155375 au
E(N2)= -109.52412868 au
E(H2)= -1.15928020 au
3*E(H2)= -3.4778406 au
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -298.2 KJ/mol
The energy for converting hydrogen and nitrogen gas into ammonia gas is -298.2 KJ/mol, which means this reaction process is exothermic, thus the ammonia product is more stable than the gaseous reactants.
NF3 optimisation
Molecule name: NF3
Calculation method: RB3LYP
Basis set: 6-31G(d.p)
Final energy E(RB3LYP) in atomic units(au): -354.07131058au
What is the RMS gradient(au)? 0.00010256 au
the point group of your molecule: C3v
optimised N-F bond distance: 1.38Å
optimised F-H-F bond angle: 102°
item table:
Item Value Threshold Converged? Maximum Force 0.000164 0.000450 YES RMS Force 0.000108 0.000300 YES Maximum Displacement 0.000612 0.001800 YES RMS Displacement 0.000296 0.001200 YES
File:NYXPHUNT NF3 OPTF POP.LOG
jmol diagram:
NF3 |
The optimisation file is liked to File:NYXPHUNT NF3 OPTF POP.LOG
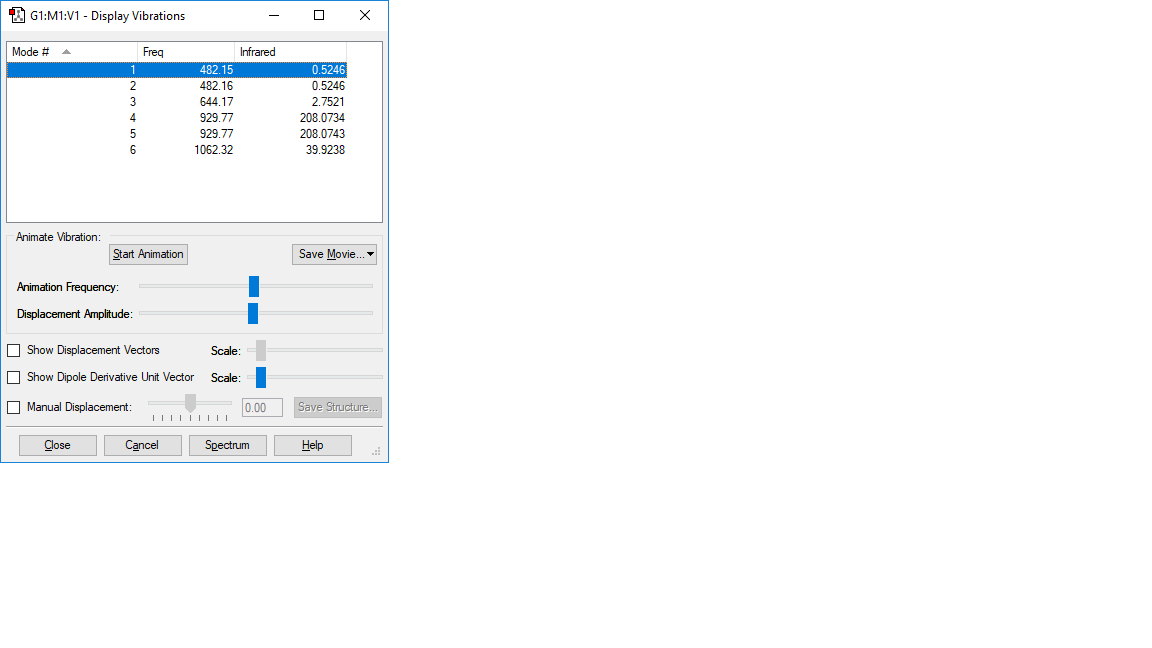
NF3 vibration:
| wavenumber cm-1 | 482 | 482 | 644 | 930 | 930 | 1062 |
|---|---|---|---|---|---|---|
| symmetry | E | E | A1 | E | E | A1 |
| intensity arbitrart units | 1 | 1 | 3 | 208 | 208 | 40 |
NBO charges:
Charge for N: 0.660, charge for F: -0.220
Molecular orbitals of NF3:
 Energy:-0.35162 au. This occupied MO is made up from the 2p AOs. It is a bonding MO, deep in energy because it is the HOMO.
Energy:-0.35162 au. This occupied MO is made up from the 2p AOs. It is a bonding MO, deep in energy because it is the HOMO.
 Energy: 0.01947 au. This unoccupied MO is made up from the 2p AOs. It is an antibonding MO, which is high in energy because it is the LUMO.
Energy: 0.01947 au. This unoccupied MO is made up from the 2p AOs. It is an antibonding MO, which is high in energy because it is the LUMO.
 Energy:-0.42224 au. This unoccupied MO is made up from four 2p orbitals of F. It is an antibonding MO, which is high in energy. 3 nodes in total.
Energy:-0.42224 au. This unoccupied MO is made up from four 2p orbitals of F. It is an antibonding MO, which is high in energy. 3 nodes in total.
 Energy:-1.23341 au. There are one 2s antibonding of F and two 2s bonding orbitals of F. One nodal plane shown.
Energy:-1.23341 au. There are one 2s antibonding of F and two 2s bonding orbitals of F. One nodal plane shown.
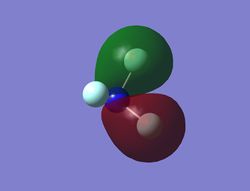 Energy:-1.23341 au. This occupied MO is made up from two s antibondings between F and N.
Energy:-1.23341 au. This occupied MO is made up from two s antibondings between F and N.
[Reference for NF3 Mo diagram]

Marking
Note: All grades and comments are provisional and subjecct to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have recieved your grade from blackboard.
Wiki structure and presentation 1/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES
NH3 0.5/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
NO
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
NO - You missed to answer the question about the atomic charges.
N2 and H2 0.5/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES, you could have explained that the charges are 0 as the electronegativities are equal.
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES
Haber-Bosch reaction energy calculation 0.5/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
NO - The energy you calculated is twice as big as the value should be. Probably there is a mistake in the calculation of ΔE
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 2/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
You have done a good job of presenting this information, well done! You should have explained the charges using an electronegativity argument. You should have analysed the calculated vibrations. You correctly displayed 5 MOs and their relative energies. However, you made mistakes labelling them as occupied or unoccupied. All MOs with a lower energy than the HOMO should be occupied! You tried to identify the AOs contributing to the MOs. However, there are several mistakes (e.g. you neglected the contribution of the N 2p orbital to the MOs 4 and 5)
Independence 0/1
If you have finished everything else and have spare time in the lab you could: Check one of your results against the literature, or Do an extra calculation on another small molecule, or Do some deeper analysis on your results so far
NO - No independent work has been identified.