Rep:Mod:nwc18
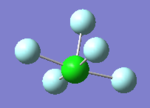
NH3 molecule
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final energy E(RB3LYP): -56.55776873 a.u.
RMS Gradient Norm: 0.00000485 a.u.
Point group: C3V
N-H bond distance: 1.02 Å accurate to 0.01 Å
H-N-H bong angle: 106 ° accurate to 1 °
Item table
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES Predicted change in Energy=-5.986284D-10
Structure of NH3
NH3 |
The optimisation file is linked to here
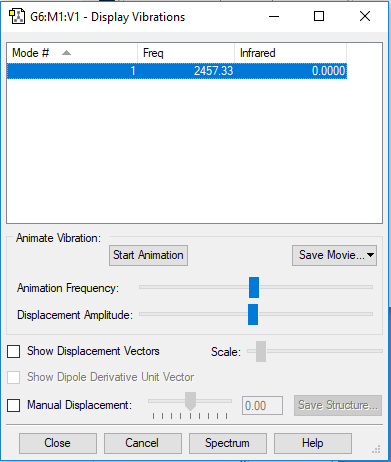
Display Vibration
Wavenumber and intensity table
| Wavenumber/cm-1 | 1090 | 1694 | 1694 | 3461 | 3590 | 3590 |
|---|---|---|---|---|---|---|
| Symmetry | A1 | E | E | A1 | E | E |
| Imtensity/arbitrary unit | 145 | 14 | 14 | 1 | 0 | 0 |
| Image |
Expect 6 modes from the 3N-6 rule.
The modes with frequency 1694 cm-1 are degenerate and the modes with frequency 3590 cm-1 are degenerate.
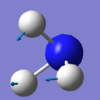
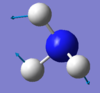
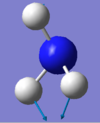
Bending vibrations: modes with frequency 1090 cm-1 and 1694 cm-1.
Bond stretch vibrations: modes with frequency 3461 cm-1 and 3590 cm-1.
The mode with frequency 3461 cm-1 is highly symmetric.
The mode with frequency 1090 cm-1 is the umbrella mode.
2 bands would be expected in an experimental spectrum of of gaseous ammonia.
Charge
Nitrogen=-1.125
Hydrogen= 0.375
The nitrogen atom is negatively charged whilst all three hydrogen atoms are positively charged because nitrogen is more electronegative than hydrogen.
N2 molecule
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final energy E(RB3LYP): -109.52412868 a.u.
RMS Gradient Norm: 0.00000060 a.u.
Point group: D∞H
N,N triple bond distance: 1.11 Å accurate to 0.01 Å
Item Table
Item Value Threshold Converged? Maximum Force 0.000001 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES Predicted change in Energy=-3.401042D-13
Structure of N2
N2 |
The optimisation file is linked to here
Display Vibration
Wavenumber and intensity table
| Wavenumbercm-1 | Symmetry | Imtensityarbitrary unit | Image |
|---|---|---|---|
| 2457 | SGG | 0 | 
|
Charge
both N atoms=0
This is because both atoms have the same electronegativity so no dipole as no difference in electronegativity.
H2 molecule
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final energy E(RB3LYP): -1.17853936 a.u.
RMS Gradient Norm: 0.00000015 a.u.
Point group: D∞H
H-H bond distance: 0.74 Å accurate to 0.01 Å
Item Table
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES Predicted change in Energy=-9.062231D-14
Structure of H2
H2 |
The optimisation file is linked to here
Display Vibration
Wavenumber and intensity table
| Wavenumbercm-1 | Symmetry | Imtensityarbitrary unit | Image |
|---|---|---|---|
| 4466 | SGG | 0 | 
|
Charge
both H atoms=0
This is because both atoms have the same electronegativity so no dipole as no difference in electronegativity.
mono-metallic TM complex that coordinates N2
unique identifier=VEJROU
link to the structure of VEJROU[[1]]
computed N,N triple bond distance=1.11 Å
experimental N,N triple bond distance in complex=1.20 Å
VEJROU is an Iron-Dinitrogen complex that catalyse the conversion of nitrogen gas into silylamine similar to the nitrogen fixation process.[1] This is essential as most nitrogen exist in the gas form in air but is required by most organisms as a nutrient. Hence, processes like nitrogen fixation allows the naturally ocurring nitrogen gas to be converted and used by organisms and so initiates the nitrogen nutrient cycle. The computational and experimental value of the N,N bond length are very similar despite the experimental value being determined from a dinitrogen metal complex other than nitrogen gas. This is because the N2 -metal interaction is weak as the central iron ion has only a charge of +2 and has a coordination number of 6. However, the small increase in bond length in the complex will be due to the positively charged iron(II) ion distorting and attracting the electron cloud of the bonding nitrogen atom and hence reducing the strength of the N,N triple bond.
Haber-Bosch reaction energy calculation
E(NH3)= -56.55777 a.u.
2*E(NH3)= -113.11554 a.u.
E(N2)= -109.52413 a.u.
E(H2)= -1.17854 a.u.
3*E(H2)= -3.53562 a.u.
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.05579 a.u.= -146.5 kJ/mol
The reaction is exothermic meaning energy was transferred to the surroundings when hydrogen and nitrogen gas react to form ammonia gas. Therefore, the ammonia product is more stable than the gaseous reactants.
ClF5 molecule
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final energy E(RB3LYP): -958.98366366 a.u.
RMS Gradient Norm: 0.00012016 a.u.
Point group: C4V
Cl-F bond distance: 1.65 Å (apical)and 1.70 Å (basal) accurate to 0.01 Å
F-Cl-F bong angle: 86 ° (apical) and 90 ° (basal) accurate to 1 °
Item Table
Item Value Threshold Converged? Maximum Force 0.000398 0.000450 YES RMS Force 0.000092 0.000300 YES Maximum Displacement 0.001404 0.001800 YES RMS Displacement 0.000374 0.001200 YES Predicted change in Energy=-3.987469D-07
Structure of ClF5
ClF5 |
The optimisation file is linked to here
Display Vibration
Wavenumber and intensity table
| Wavenumber/cm-1 | 259 | 280 | 280 | 374 | 452 | 452 | 471 | 522 | 560 | 733 | 805 | 805 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Symmetry | B2 | E | E | B1 | E | E | A1 | B2 | A1 | A1 | E | E |
| Imtensity/arbitrary unit | 0 | 0 | 0 | 0 | 0 | 0 | 28 | 0 | 7 | 75 | 404 | 404 |
| Image |
All B symmetries are not IR active because there is no dipole moment. This is because the vibration of the flourine molecules allows the dipoles to cancel out.
Expect 12 modes from the 3N-6 rule.
The modes with frequency 280 cm-1, 452cm-1and 805cm-1 are degenerate with each of the same frequency..
Bending vibrations: modes with frequency 259 cm-1, 280 cm-1, 347 cm-1,452 cm-1 and 471 cm-1.
Bond stretch vibrations: modes with frequency 522 cm-1, 560 cm-1, 733 cm-1 and 805 cm-1.
The mode with frequency 560 cm-1 and 733 cm-1 are highly symmetric.
4 bands would be expected in an experimental spectrum of of gaseous ammonia.
Charge
Chlorine= 1.998
Fluorine= -0.335 (apical) and -0.416 (basal)
Fluorine atoms are negatively charged whilst chlorine atoms are poistively charged because fluorine is more electronegative (the most electronegative) than chlorine.
Molecular Orbital
Hybridisation of the chlorine atom in ClF5 = sp3d2
The molecular structure of ClF5 = square pyramidal
This structure is due the lone pair of electrons on the chlorine atom repelling the 5 singly bonded fluorine atoms more than the bonding pairs repel each other.
The apical bond and basal bonds are similar in bond energy and so bond length. This is because the bond angle between two opposite basal bonds is 171 ° which is close to the crossover point where the trend in bond energy reverse.[2]
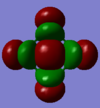
| MO 1.σ
(lowest molecular orbital) |
MO 2.σ*
(valence bond orbital) |
MO 3.σ*
(valence bond orbital) |
MO 4.σ*Homo
(valence bond orbital) |
MO 5.σ*Lumo |
|---|---|---|---|---|
|

|

|
|

|
MO 1.σ:The overlap of the 1s orbitals cannot be seen as the orbitals are too small and low in energy(-101.96235 a.u.). Hence, this will not be involved in bonding.
MO 2 and 3.σ*: The p orbitals overlap to form both σ and π bonds. There are three p orbitals on the fluorine atoms which contributes to the σ* MO but the 3pz orbital has the greatest contribution to the σ* MO. This is because σ antibonding orbitals are higher in energy than bonding orbitals and π antibonding orbitals in the same set due to the lower density of electron between the nuclei. The two σ* (MO 2 and 3) are degenerate.
HOMO: This is an anitbonding MO with the s orbital of chlorine and p orbitals of the fluorines being the main contributor. This is the highest occupied molecular orbital invovled in bonding.
LUMO: This is a σ* orbital with both antibonding and bonding characters. The py orbitals on the basal fluorines and the pz orbitals of the chlorine and apacal fluorine are the main contributions towards the MO. The pz orbitals of the apacal fluorine and chlorine are antibonding whilst the py orbitals of the basal fluorine shows bonding character towards the pz orbital of the chlorine (as shown in the LUMO MO diagram). This is not involved in bonding.
Independent mark:O2 molecule
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final energy E(RB3LYP): -150.25742434 a.u.
RMS Gradient Norm: 0.00007502 a.u.
Point group: D∞H
O-O bond distance: 1.22 Å accurate to 0.01 Å
Item Table
Item Value Threshold Converged? Maximum Force 0.000130 0.000450 YES RMS Force 0.000130 0.000300 YES Maximum Displacement 0.000080 0.001800 YES RMS Displacement 0.000113 0.001200 YES Predicted change in Energy=-1.033738D-08
Structure of O2
O2 |
The optimisation file is linked to here
Display Vibration
Wavenumber and intensity table
| Wavenumbercm-1 | Symmetry | Imtensityarbitrary unit | Image |
|---|---|---|---|
| 1643 | SGG | 0 |
Charge
both O atoms=0
This is because both atoms have the same electronegativity so no dipole as no difference in electronegativity.
References
- ↑ Imayoshi,R.,Nakajima,K.,Takaya,J.,Iwasawa,N.,Nishibayashi,Y.(2017) Synthesis and Reactivity of Iron- and Cobalt-Dinitrogen Complexes Bearing PSiP-Type Pincer Ligands toward Nitrogen Fixation. European Journal of Inorganic Chemistry. 2017(32),3769-3778. Available from:https://doi.org/10.1002/ejic.201700569 [Accessed 19th March 2019].
- ↑ Rossi,A,R.,Hoffmann,R.(1974) Transition Metal Pentacoordination. Inorganic Chemistry. 1975,14(2),365-374. Available from:https://pubs.acs.org/doi/pdf/10.1021/ic50144a032 [Accessed 21th March 2019].
Marking
Note: All grades and comments are provisional and subject to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have received your grade from blackboard.
Wiki structure and presentation 1/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES
NH3 1/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES
N2 and H2 0.5/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES
Haber-Bosch reaction energy calculation 1/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 3.5/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
YES, your charges explanation is good and you have made a good attempt at explaining the MOs (which are quite complicated for this molecule!). For a complicated molecule like this it is often not possible to label an entire MO as sigma or pi, or even bonding or antibonding. Instead you can talk about the individual interactions present for example in MO2 you could say that there are sigma type antibonding interactions between the neighbouring F atoms, and that this MO reduces the F-F bonding character.
Independence 1/1
If you have finished everything else and have spare time in the lab you could:
Check one of your results against the literature, or
Do an extra calculation on another small molecule, or
Do some deeper analysis on your results so far
You did an extra calculation on O2 well done!