Rep:Mod:ns618
Optimzed NH3
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986278D-10
Optimization completed.
-- Stationary point found.
| Molecule | NH3 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final Energy E(RB3LYP) | -56.55776873 a.u. |
| RMS Gradient | 0.00000485 a.u. |
| Point Group | C3V |
N-H Bond Length : 1.01798 angstrom
H-N-H Bond Angle : 105.741 degrees
optimized NH3 |
| Wavenumber (/cm) | 1089.5366 | 1693.9474 |
| Symmetry | A1 | E |
| intensity (arbitrary units) | 145.3814 | 13.5533 |
Charge on N-atom = -1.125 Charge on H-atom = +0.375 Expected charge on N-atom is -3 and on H-atom is +1
Optimzed H2
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
| Molecule | H2 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final Energy E(RB3LYP) | -1.17853936 a.u. |
| RMS Gradient | 0.00000017 a.u. |
| Point Group | D*H |
H-H Bond Length : 0.74279 angstrom
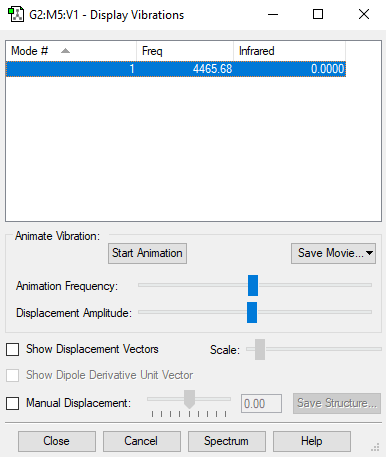
optimized NH3 |
| Wavenumber (/cm) | 4465.6824 |
| Symmetry | SGG |
| intensity (arbitrary units) | 0 |
Charge on H-atom = 0 Expected charge on H-atom is 0
Optimzed N2
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.383850D-13
Optimization completed.
-- Stationary point found.
| Molecule | N2 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final Energy E(RB3LYP) | -109.52412868 a.u. |
| RMS Gradient | 0.00000060 a.u. |
| Point Group | D*H |
H-H Bond Length : 1.10550 angstrom
optimized NH3 |
| Wavenumber (/cm) | 2457.3283 |
| Symmetry | SGG |
| intensity (arbitrary units) | 0 |
Charge on N-atom = 0 Expected charge on N-atom is 0
Optimzed P4
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000017 0.001800 YES
RMS Displacement 0.000013 0.001200 YES
Predicted change in Energy=-4.333417D-11
Optimization completed.
-- Stationary point found.
| Molecule | P4 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final Energy E(RB3LYP) | -1365.33522654 a.u. |
| RMS Gradient | 0.00000144 a.u. |
| Point Group | C1 |
P-P Bond Length : 1.95272, 2.18213, 2.10909 angstrom
P-P-P Bond Angle : 151.102, 61.101, 57.797 degrees
optimized NH3 |
| Wavenumber (/cm) | 87.9479 | 152.7457 | 337.3398 |
| Symmetry | A | A | A |
| intensity (arbitrary units) | 0.1973 | 0.1116 | 4.0377 |
Charge on P-atom = -0.129, -0.30, +0.79, +0.79 Expected charge on P-atom is 0
Marking
Note: All grades and comments are provisional and subject to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have received your grade from blackboard.
Wiki structure and presentation 0.5/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
NO - you have not used headings at all in the way that a table of contents was generated. All your jmols are labelled as 'optimised NH3" which is only true for one of them.
NH3 0/1
Have you completed the calculation and given a link to the file?
NO - You missed to include a link to the .log file of your finished calculation. This reduces the achievable mark for this section by 1.
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES - but it is missing most of the vibrations.
Did you do the optional extra of adding images of the vibrations?
NO
Have you included answers to the questions about vibrations and charges in the lab script?
NO - you missed to answer all questions regarding the vibrations.
N2 and H2 0/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
NO - You missed to include a link to the .log file of your finished calculation. This reduces the achievable mark for this section by 1.
Crystal structure comparison 0/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
NO
Have you compared your optimised bond distance to the crystal structure bond distance?
NO
Haber-Bosch reaction energy calculation 0/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
NO
Have you reported your answers to the correct number of decimal places?
NO
Do your energies have the correct +/- sign?
NO
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
NO
Your choice of small molecule 0.5/5
Have you completed the calculation and included all relevant information?
NO - You missed to include a link to the .log file of your finished calculation. This reduces the achievable mark for this section by 1.
Have you added information about MOs and charges on atoms?
You only stated information about and vibrations without trying to analyse them. You stated the charges of the P atoms. You only stated the charge you would have expected but gave no comparison or explanation why the computed charges are different. This is probably because the geometry of your P4 molecule is not looking like it is in reality. Yours is planar but it should be a tetrahedron.
Independence 0/1
If you have finished everything else and have spare time in the lab you could:
Check one of your results against the literature, or
Do an extra calculation on another small molecule, or
Do some deeper analysis on your results so far
NO - No independent work has been identified.
Moderation
The marking of this wiki has been moderated by Tricia Hunt and the grade confirmed. You did complete a few tasks, and we hope the lab was useful/enjoyable to you and taught you something about computational chemistry!
A note on your grade: Unfortunately in this case the pieces of work were thinly spread across different sections of the lab, so in most cases there was simply too little work present in each section to achieve the minimum standard required to award any marks. A similar level of work applied to just one section of the mark scheme may well have resulted in more marks.