Rep:Mod:newton
James Cochrane's Wiki
BH3
3D View Of Optimised BH3
Smf115 (talk) 23:51, 16 May 2018 (BST)Very nice structure information and presentation throughout, good addition of the geomotries on the jmols!
Calculation Results For Optimised BH3
| Summary Table | |
|---|---|
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Energy | -26.61532 a.u. |
| RMS Gradient Norm | 0.00008 a.u. |
| Dipole Moment | 0.00 Debye |
| Point Group | CS |
| Calculation Time | 3 min 1 s |
| Item Table | |
Item Value Threshold Converged? Maximum Force 0.000203 0.000450 YES RMS Force 0.000098 0.000300 YES Maximum Displacement 0.000867 0.001800 YES RMS Displacement 0.000415 0.001200 YES | |
| Optimised Parameters | |
Optimized Parameters
(Angstroms and Degrees)
Name Definition Value Derivative Info.
R1 R(1,2) 1.1924 -DE/DX = 0.0
R2 R(1,3) 1.1928 -DE/DX = -0.0002
R3 R(1,4) 1.1926 -DE/DX = -0.0002
A1 A(2,1,3) 119.9864 -DE/DX = 0.0
A2 A(2,1,4) 119.999 -DE/DX = 0.0
A3 A(3,1,4) 120.0146 -DE/DX = 0.0
D1 D(2,1,4,3) 180.0 -DE/DX = 0.0
| |
| BH3 optimisation *.log file | |
Frequency Analysis
Low Frequencies
Low frequencies -0.2268 -0.0080 0.0008 22.1147 22.1159 24.1363 Low frequencies 1163.1747 1213.2735 1213.2737
Vibrations
| Intensity (arbitrary units) | Irreducible representation | IR active? | Vibration Type | |
|---|---|---|---|---|
| 1163 | 93 | A2 | ✓ | Wagging |
| 1213 | 14 | E | Slightly | In-plane scissoring |
| 1213 | 14 | E | Slightly | In-plane stretch |
| 2582 | 0 | A | ✗ | In-plane symmetric stretch |
| 2715 | 126 | E | ✓ | Antisymmetric stretch |
| 2715 | 126 | E | ✓ | Symmetric stretch |
Calculated IR spectrum

We expected to see 6 vibrations from the 3N-6 rule for non-linear molecules. However, we only observe 3 in the spectrum. Peak at 2582 cm-1 has zero intensity because the vibration is an in-plane, fully symmetric stretch. The degenerate pair of peaks at 1213 cm-1 should have zero intensity because the E irreducible representation doesn't have the same symmetry species as the displacements x,y or z (see D3h character table). However, because the optimised structure of BH3 is not completely symmetric, the E vibrations can non-zero intensity.
Molecular Orbitals
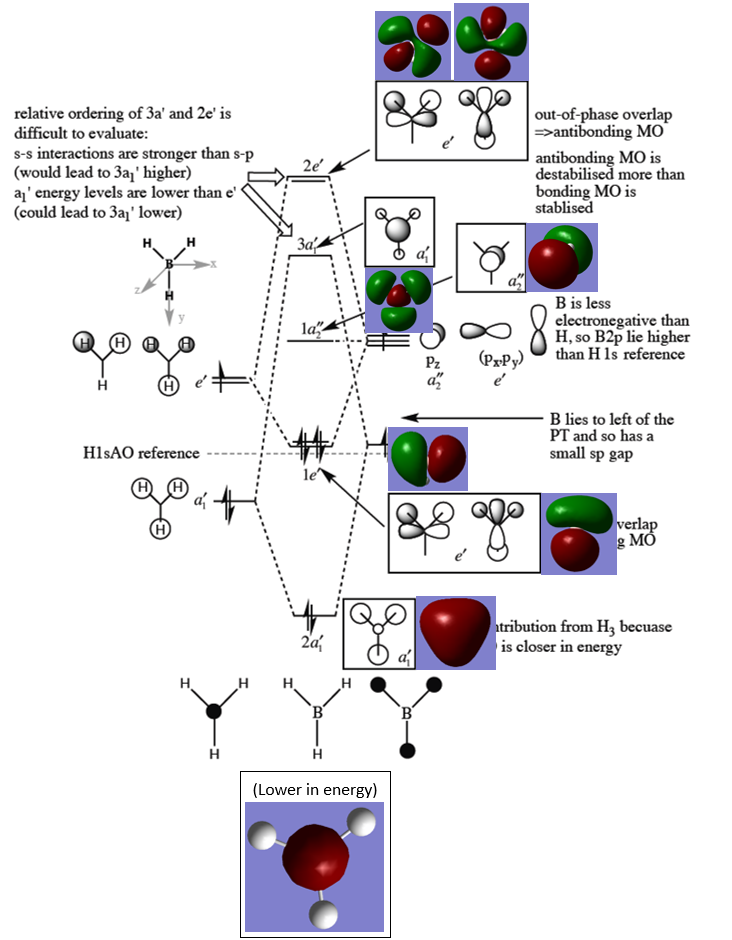
The shapes and phases of the LCAO MOs are accurate for the lower 5 MOs. However, the shapes of the 3 highest energy LCAO MOs are slightly different to the computed MO shapes. Despite this, qualitative MO theory perfectly predicts orbital degeneracies and provides a useful, accurate way of determining orbital shapes for BH3.
Smf115 (talk) 23:51, 16 May 2018 (BST)Good annotation of the MO diagram and mention to both the similarities and differences.
Association Energy
NH3
3D View Of Optimised NH3
Calculation Results For Optimised NH3
| Summary Table | |
|---|---|
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Energy | -56.55777 a.u. |
| RMS Gradient Norm | 0.00000 a.u. |
| Dipole Moment | 1.85 Debye |
| Point Group | C3v |
| Calculation Time | 2 min 2 s |
| Item Table | |
Item Value Threshold Converged? Maximum Force 0.000006 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000012 0.001800 YES RMS Displacement 0.000008 0.001200 YES | |
| Optimised Parameters | |
Optimized Parameters
(Angstroms and Degrees)
Name Definition Value Derivative Info.
R1 R(1,2) 1.018 -DE/DX = 0.0
R2 R(1,3) 1.018 -DE/DX = 0.0
R3 R(1,4) 1.018 -DE/DX = 0.0
A1 A(2,1,3) 105.7446 -DE/DX = 0.0
A2 A(2,1,4) 105.7446 -DE/DX = 0.0
A3 A(3,1,4) 105.7446 -DE/DX = 0.0
D1 D(2,1,4,3) -111.8637 -DE/DX = 0.0
| |
| NH3 optimisation *.log file | |
Frequency Analysis
Low Frequencies
Low frequencies -0.0138 -0.0032 -0.0015 7.0783 8.0932 8.0937 Low frequencies 1089.3840 1693.9368 1693.9368
NH3BH3
3D View Of Optimised NH3BH3
Calculation Results For Optimised NH3BH3
| Summary Table | |
|---|---|
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Energy | -83.22469 a.u. |
| RMS Gradient Norm | 0.00006 a.u. |
| Dipole Moment | 5.57 Debye |
| Point Group | C1 |
| Calculation Time | 2 min 48 s |
| Item Table | |
Item Value Threshold Converged? Maximum Force 0.000123 0.000450 YES RMS Force 0.000058 0.000300 YES Maximum Displacement 0.000515 0.001800 YES RMS Displacement 0.000296 0.001200 YES | |
| Optimised Parameters | |
Optimized Parameters
(Angstroms and Degrees)
Name Definition Value Derivative Info.
R1 R(1,7) 1.0186 -DE/DX = -0.0001
R2 R(2,7) 1.0186 -DE/DX = -0.0001
R3 R(3,7) 1.0186 -DE/DX = -0.0001
R4 R(4,8) 1.21 -DE/DX = -0.0001
R5 R(5,8) 1.21 -DE/DX = -0.0001
R6 R(6,8) 1.21 -DE/DX = -0.0001
R7 R(7,8) 1.6681 -DE/DX = -0.0001
A1 A(1,7,2) 107.8687 -DE/DX = 0.0
A2 A(1,7,3) 107.8687 -DE/DX = 0.0
A3 A(1,7,8) 111.0301 -DE/DX = 0.0
A4 A(2,7,3) 107.8686 -DE/DX = 0.0
A5 A(2,7,8) 111.0296 -DE/DX = 0.0
A6 A(3,7,8) 111.0297 -DE/DX = 0.0
A7 A(4,8,5) 113.8744 -DE/DX = 0.0
A8 A(4,8,6) 113.8743 -DE/DX = 0.0
A9 A(4,8,7) 104.5966 -DE/DX = 0.0
A10 A(5,8,6) 113.8744 -DE/DX = 0.0
A11 A(5,8,7) 104.5969 -DE/DX = 0.0
A12 A(6,8,7) 104.597 -DE/DX = 0.0
D1 D(1,7,8,4) 179.9996 -DE/DX = 0.0
D2 D(1,7,8,5) -60.0004 -DE/DX = 0.0
D3 D(1,7,8,6) 59.9997 -DE/DX = 0.0
D4 D(2,7,8,4) -60.0002 -DE/DX = 0.0
D5 D(2,7,8,5) 59.9997 -DE/DX = 0.0
D6 D(2,7,8,6) 179.9999 -DE/DX = 0.0
D7 D(3,7,8,4) 59.9995 -DE/DX = 0.0
D8 D(3,7,8,5) 179.9995 -DE/DX = 0.0
D9 D(3,7,8,6) -60.0004 -DE/DX = 0.0
| |
| NH3BH3 optimisation *.log file | |
Frequency Analysis
Low Frequencies
Low frequencies -0.0251 -0.0030 0.0012 17.1236 17.1258 37.1326 Low frequencies 265.7816 632.2034 639.3483
Energy Calculation
| Molecule | E(RB3LYP)/Ha |
|---|---|
| BH3 | -26.61532 |
| NH3 | -56.55777 |
| NH3BH3 | -83.22469 |
The dissociation energy is therefore (E(NH3) + E(BH3)) - E(NH3BH3) = - 26.61532 - 56.55777 + 83.22469 Ha. Since Ha = 2625.499638(65) kJ/mol, the dissociation energy = + 135 kJ/mol. This value is accurate when compared to the literature value of 145 kJ/mol.[2] When this bond dissociation energy is compared to that of the covalent CH3-H bond, 423 kJ/mol,[3] it is clear that the B-N bond is much weaker than a covalent bond. However, when compared to the hydrogen bond strength for a water-neon molecule pair, 0.77 kJ/mol,[4] the B-N bond is a medium strength dative bond.
Smf115 (talk) 22:13, 15 May 2018 (BST)Really thorough and well referenced answer and calculation. Good attention to the accuracy of the figures recorded.
BBr3
3D View Of Optimised BBr3
Calculation Results For Optimised BBr3
| Summary Table | |
|---|---|
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | Gen |
| Energy | -64.43645 a.u. |
| RMS Gradient Norm | 0.00001 a.u. |
| Dipole Moment | 0.00 Debye |
| Point Group | CS |
| Calculation Time | 0 min 35 s |
| Item Table | |
Item Value Threshold Converged? Maximum Force 0.000011 0.000450 YES RMS Force 0.000007 0.000300 YES Maximum Displacement 0.000047 0.001800 YES RMS Displacement 0.000031 0.001200 YES | |
| Optimised Parameters | |
Optimized Parameters
(Angstroms and Degrees)
Name Definition Value Derivative Info.
R1 R(1,2) 1.934 -DE/DX = 0.0
R2 R(1,3) 1.934 -DE/DX = 0.0
R3 R(1,4) 1.9339 -DE/DX = 0.0
A1 A(2,1,3) 119.9973 -DE/DX = 0.0
A2 A(2,1,4) 120.0028 -DE/DX = 0.0
A3 A(3,1,4) 119.9999 -DE/DX = 0.0
D1 D(2,1,4,3) 180.0 -DE/DX = 0.0
| |
| BBr3 optimisation *.log file | |
Frequency Analysis
Low Frequencies
Low frequencies -0.0001 0.0002 0.0002 0.6823 1.2779 2.4660 Low frequencies 155.9396 155.9490 267.6878
BBr3 frequency B3LYP/6-31G(d,p)LANL2DZ DSpace link
Ionic Liquids: Designer Solvents
NMe4+
3D View Of Optimised NMe4+
Calculation Results For Optimised NMe4+
| Summary Table | |
|---|---|
| File Type | .log |
| Calculation Type | Opt+Freq |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Energy | -214.18127 a.u. |
| RMS Gradient Norm | 0.00007 a.u. |
| Dipole Moment | 0.00 Debye |
| Point Group | C1 |
| Charge | 1 |
| Calculation Time | 4 min 5 s |
| Item Table | |
Item Value Threshold Converged? Maximum Force 0.000249 0.000450 YES RMS Force 0.000036 0.000300 YES Maximum Displacement 0.000853 0.001800 YES RMS Displacement 0.000253 0.001200 YES | |
| Optimised Distances | |
Optimized Parameters
(Angstroms)
Name Definition Value Derivative Info.
R1 R(1,2) 1.0901 -DE/DX = 0.0
R2 R(1,3) 1.0901 -DE/DX = 0.0
R3 R(1,4) 1.0902 -DE/DX = 0.0
R4 R(1,13) 1.5095 -DE/DX = 0.0
R5 R(5,7) 1.0901 -DE/DX = 0.0
R6 R(5,8) 1.0902 -DE/DX = 0.0
R7 R(5,9) 1.0901 -DE/DX = 0.0
R8 R(5,13) 1.5095 -DE/DX = 0.0
R9 R(6,10) 1.0902 -DE/DX = 0.0
R10 R(6,11) 1.0901 -DE/DX = 0.0
R11 R(6,12) 1.0901 -DE/DX = 0.0
R12 R(6,13) 1.5095 -DE/DX = 0.0
R13 R(13,14) 1.5088 -DE/DX = 0.0002
R14 R(14,15) 1.0903 -DE/DX = -0.0001
R15 R(14,16) 1.0903 -DE/DX = -0.0001
R16 R(14,17) 1.0903 -DE/DX = -0.0001
| |
| NMe4+ optimisation *.log file | |
Frequency Analysis
Low Frequencies
Low frequencies -12.9787 -9.0238 -4.8995 0.0005 0.0007 0.0010 Low frequencies 182.1725 287.2913 287.9973
Charge Distribution
| Atom | Charge |
|---|---|
| H | +0.269 |
| C | -0.483 |
| N | -0.295 |
The total charge on the NMe4+ is equally distributed among the four methyl sites.[5] The traditional picture of NMe4+ would localize a "formal" +1 charge on nitrogen for convenience - a reflection of the electron count associated with the isolated neutral atom compared to the atom in the molecule of concern. This computational result, which assigns a negative NBO charge on nitrogen, differs from the traditional model. Additionaly, the positive charge actually resides on the hydrogen atoms - invalidating the traditional picture.
Molecular Orbitals
Using this representation of the fragment orbitals, it is clear to see the fragment orbitals of MO11, MO19 and MO10 below.
Although there is a weak "in-phase" interaction between the three "p-like" methyl fragment orbitals and the fourth "s-like" orbital in MO19, MO19 possesses more nodes than MO11. It's greater antibonding character means that MO19 is higher in energy than MO11.
MO10 is highly symmetric because it comprises 4, "s-like" methyl fragment orbitals and an "s" orbital on nitrogen of opposite phase. It is lower in energy than MO11 and MO19 because there is much greater total "s" character compared to the "p" character of MO11/MO19. The shapes of the LCAO MOs accurately predict the true MO shapes for MO19, MO11 and MO10.
Smf115 (talk) 23:44, 16 May 2018 (BST)Correct LCAO approach showing understanding of how to derive MOs this way. However, a better approach would be to choose the MO here and then to break it down in to it's fragments as the ones show here aren't fully correct. Consider the BH3 MOs seen in your lectures and how these relate to the CH3 ligand FO's.
PMe4+
3D View Of Optimised PMe4+
Calculation Results For Optimised PMe4+
| Summary Table | |
|---|---|
| File Type | .log |
| Calculation Type | Opt+Freq |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Energy | -500.82701 a.u. |
| RMS Gradient Norm | 0.00004 a.u. |
| Dipole Moment | 0.00 Debye |
| Point Group | C1 |
| Charge | 1 |
| Calculation Time | 4 min 7 s |
| Item Table | |
Item Value Threshold Converged? Maximum Force 0.000068 0.000450 YES RMS Force 0.000023 0.000300 YES Maximum Displacement 0.001006 0.001800 YES RMS Displacement 0.000350 0.001200 YES | |
| Optimised Distances | |
Optimized Parameters
(Angstroms)
Name Definition Value Derivative Info.
R1 R(1,2) 1.0933 -DE/DX = 0.0
R2 R(1,3) 1.0933 -DE/DX = 0.0
R3 R(1,4) 1.0933 -DE/DX = 0.0
R4 R(1,13) 1.8165 -DE/DX = 0.0
R5 R(5,7) 1.0933 -DE/DX = 0.0
R6 R(5,8) 1.0933 -DE/DX = 0.0
R7 R(5,9) 1.0933 -DE/DX = 0.0
R8 R(5,13) 1.8166 -DE/DX = -0.0001
R9 R(6,10) 1.0933 -DE/DX = 0.0
R10 R(6,11) 1.0933 -DE/DX = 0.0
R11 R(6,12) 1.0933 -DE/DX = 0.0
R12 R(6,13) 1.8166 -DE/DX = 0.0
R13 R(13,14) 1.8165 -DE/DX = 0.0
R14 R(14,15) 1.0933 -DE/DX = 0.0
R15 R(14,16) 1.0933 -DE/DX = 0.0
R16 R(14,17) 1.0933 -DE/DX = 0.0
| |
| PMe4+ optimisation *.log file | |
Frequency Analysis
Low Frequencies
Low frequencies -7.0674 -0.0028 -0.0017 -0.0012 3.8043 6.4753 Low frequencies 155.9547 191.2644 191.9015
Charge Distribution
| Atom | Charge |
|---|---|
| H | +0.298 |
| C | -1.060 |
| P | +1.667 |
In PMe4+ the phosphorus atom accommodates more positive charge than the nitrogen atom does in NMe4+ (+1.667 vs -0.295). This can be explained by the electronegativities of C,N and P - 2.55, 3.04 and 2.19 (Pauling scale) respectively. N is the most electronegative atom so its negative calculated NBO charge is reasonable. On the other hand, P is the least electronegative atom so its significant positive NBO charge is fair.
Smf115 (talk) 23:40, 16 May 2018 (BST)Good explaination of the charges due to the electronegativities, however, comparison of the C and H charges across the molecules would have improved the discussion.
Smf115 (talk) 23:53, 16 May 2018 (BST)Overall a well presented wiki report with a strong first section and further consideration of the MOs needed in the second section.
References
- ↑ Hunt Research Group, http://www.huntresearchgroup.org.uk/teaching/teaching_comp_lab_year2a/Tut_MO_diagram_BH3.pdf, (accessed May 2018).
- ↑ J. S. Binkley and L. R. Thorne, J. Chem. Phys., 1983, 79, 2932.
- ↑ B. E. Knox and H. B. Palmer, Chem. Rev. (Washington, DC, U. S.), 1961, 61, 247-255.
- ↑ M. Losonczy and J. W. Moskowitz, J. Chem. Phys., 1974, 61, 2438.
- ↑ S. Garde, G. Hummer and M. E. Paulaitis, J. Chem. Phys., 1998, 108, 1553.



