Rep:Mod:musiq-child(remixed)
Module 2: Bonding (Ab Initio and Density Functional MO Methods)
Here the Gaussian (09W) and Gaussview (5.0) programs on the laptop will be used (alongside the Imperial Chemistry SCAN) to determine the optimised geometries and essential properties of these systems (structure, bonding, and more specifically absolute energies (a.u.), relative energies (kJ mol-1), molecular orbitals, Natural Bond Orbital analysis results (charges on atoms, polarity/contributions to bonds), and frequencies of key vibrations in the molecules (cm-1). Interpretation of these results and key discussion will follow.
Before beginning any molecular analysis, a folder was created to contain all files: H:\Comp. Labs\Mod 2.
Basic Techniques of Optimisation and Analysis: BH3 and BCl3
BH3
A molecule of BH3 was created in the molecule window by selecting B from the periodic table function in the palette window-the planar fragment was chosen-and clicking once in the molecule window. In Inquiry mode, the molecule was manipulated to get used to this, e.g. translation, magnify/shrink, and rotate (X,Y,Z-directions), and the atomic symbols were displayed. Use of the Fragment menus was investigated as well in a separate molecule window.
For pre-optimisation purposes, the bond length/angle/dihedral angle parameters were also investigated in a separate molecule window, before the B-H bond lengths were all changed to 1.5Å on our main BH3 molecule.
The geometry of BH3 was optimised using:
Method: DFT-B3LYP; Basis set: 3-21G; Calculation: OPT
where these parameters were entered in the Gaussian Calculation Setup tab, and the title given was "BH3 Optimisation". The file was saved with name "hiren_bh3_opt" (no spaces), thus creating the input GJF file for the calculation with these specific parameters. The "job" was then submitted to the laptop Gaussian program and run accordingly. [1]
Once the job was completed, the results .log/.out file was opened (making sure the file type is for Gaussian Output files), opening the optimised BH3 in a new (careful!) molecule window:

|
|---|
The Inquiry button was used to find the optimised bond lengths and angles: B-H bond length = 1.19Å H-B-H bond angle = 120.0°
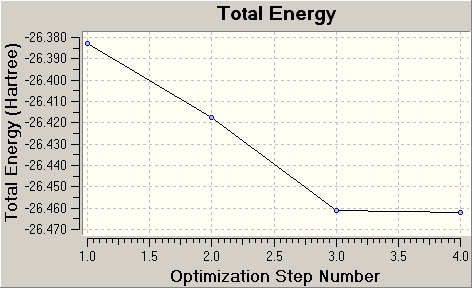
The Summary section of the Gaussian calculation was looked at, and to check that the optimisation was complete, the Gradient was looked at; as it was less than 0.001 (0.00006014) a.u., the optimisation was confirmed successful. The actual output .log/.out file was then looked at, and it was confirmed that the optimisation had converged to a maximum (or minimum; see later) by seeing that the forces and displacements had all converged (noted by a YES response to both forces and displacements). Then checking the Results, Optimisation tabs, the following graphs were given:
Here the total energy of the molecule and the RMS gradient steadily decreases as the optimisation step number increases. The gradient is the gradient of the energy profile/potential energy surface of the molecule (as a function of the atomic positions relative to each other). As there is a net force displacing these equilibrium positions, the energy changes, the atomic positions change, and so the gradient (or net force) becomes non-zero. At equilibrium this gradient is zero, hence the molecule is in its equilibrium or lowest energy geometry minimum. Hence a zero gradient result of the optimisation is desired, and this is more or less achieved by the final optimisation step 4. This is also reflected in the lowest energy value of the geometry at the 4th optimisation step. Below are the optimisation steps (showing intermediate geometries):
| Optimisation Step 1 | Optimisation Step 2 | Optimisation Step 3 | Optimisation Step 4 |
|---|---|---|---|
 |
 |
 |
 |
Looking at the first two images (steps 1 and 2), there are no bonds visualised between the atoms. This is due to usually two reasons: either the bond length is pre-defined between two particular atoms, so if this is exceeded (e.g. due to changing the bond lengths like before), then Gaussview won't draw them in; or there is no pre-defined bond length between the two atoms (usually beacuse for example coordination bonds from ligand to transition metal centres). Either way, they are still there, just Gaussview can't visualise them. The chemical bond is still there: an interaction between atoms or molecules that leads to polyatomic compounds; they can be electrostatic (attractive) interactions, or dipole interactions; the electrostatics can be between ions, or electrons between nuclei pulling them both closer to each other (i.e. covalent bond).
Following geometry optimisation of the BH3 molecule, anaylsis was performed. Firstly, MO analysis was done by opening the .chk file produced alongside the .out file from the optimisation, and re-submitting it as a GJF file with the following parameters:
Method: DFT-B3LYP; Basis Set: 3-21G; Calculation: Energy; Additional Keywords: pop=full; NBO Tab: Full NBO
The job was run with filename "hiren_bh3_pop" by SCAN, and the results were published (deposited in the chemical database "D-space"): DOI:10042/to-3797
Unable to write-up any more due to lack of time, below are all jobs run-all were successful/proved the optimised geometry was an energy minimum
Frequency Analysis of BH3: DOI:10042/to-3847
BCl3 optimisation: https://www.ch.ic.ac.uk/wiki/images/d/d2/HIREN_BCL3_OPT.LOG
BCl3 Frequency Analysis: DOI:10042/to-3876
Isomers of Mo(CO)4L2
Mo cis initial optimisation (LANL2MB): DOI:10042/to-3923
Mo trans initial optimisation (LANL2MB): DOI:10042/to-3925
Mo cis final optimisation (LANL2DZ): DOI:10042/to-4024
Mo trans final optimisation (LANL2DZ): DOI:10042/to-4025
Mo cis frequency: DOI:10042/to-4081
Mo trans frequency: DOI:10042/to-4082
MiniProject: Investigating Ammonia-Borane and derivatives
BH3-NH3 optimisation: DOI:10042/to-4044
BH3-NH3 frequency: DOI:10042/to-4083
BH3-N(CH3)3 optimisation: DOI:10042/to-4045
BH3-N(CH3)3 frequency: DOI:10042/to-4084
BH3-NF3 optimisation: DOI:10042/to-4047
BH3-NF3 frequency: DOI:10042/to-4085
BH3-P(CH3)3 optimisation: DOI:10042/to-4048
BH3-P(CH3)3 frequency: DOI:10042/to-4086