Rep:Mod:mm1213
Wiki page Computational Chemistry
Michelle Madin 01340133
NH3 molecule
Optimized N-H bond distance = 1.01798 Å
Optimized H-N-H bond angle = 105.741 °
Calculation method = RB3LYP
Basis set = 6-31G(d.p)
Final energy E(RB3LYP) = -56.55776873 a.u.
RMS gradient norm = 0.00000485 a.u.
Point group = C3V
Final set of forces and displacements for NH3 Optimization
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES
Jmol and File Link
NH3 Optimisation |
NH3 Vibration Frequencies
| Mode # | Frequency | Infrared |
|---|---|---|
| 1 | 1089.54 | 145.3814 |
| 2 | 1693.95 | 13.5533 |
| 3 | 1693.95 | 13.5533 |
| 4 | 3461.29 | 1.0608 |
| 5 | 3589.82 | 0.2711 |
| 6 | 3589.82 | 0.2711 |
How many modes do you expect from the 3N-6 rule? = 6 modes of vibration.
Which modes are degenerate? = 2 and 3 are degenerate and 5 and 6 are degenerate.
Which modes are "bending" vibrations and which are "bond stretch" vibrations? = 1,2,3 are bending and 4,5,6 are bond stretches.
Which mode is highly symmetric? = 4
Which mode is known as the "umbrella" mode? = 1
How many bands would you expect to see in an experimental spectrum of gaseous ammonia? = You would see two peaks in the spectrum , with the most intense vibration being the first. Vibrations 2 and 3 would show as a single peak due to these modes being degenerate. Vibrations 4,5 and 6 are negligible as a result of the noise in the experimental results.
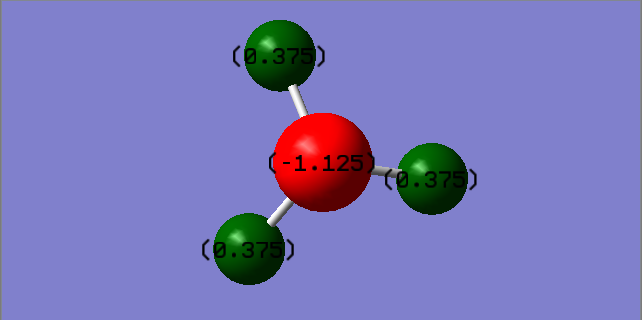
Charge Distribution of NH3
Nitrogen has the higher electronegativity and therefore has the higher electron density. This will result in nitrogen having a negative charge and the hydrogens having the positive charges, which is confirmed by the charge distribution shown above.
N2 molecule
Optimized N-N bond distance = 1.10550 Å
Optimized N-N bond angle = 180.000 °
Calculation method = RB3LYP
Basis set = 6-31G(d.p)
Final energy E(RB3LYP) = -109.52412868 a.u.
RMS gradient norm = 0.00000060 a.u.
Point group = D∞H
Final set of forces and displacements for N2 Optimization
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Jmol and File Link
N2 Optimisation |
N2 Vibration Frequencies
| Mode # | Frequency | Infrared |
|---|---|---|
| 1 | 2457.33 | 0.0000 |
H2 molecule
Optimized H-H bond distance = 0.74279 Å
Optimized H-H bond angle = 180.000 °
Calculation method = RB3LYP
Basis set = 6-31G(d.p)
Final energy E(RB3LYP) = -1.17853936 a.u.
RMS gradient norm = 0.00000017 a.u.
Point group = D∞H
Final set of forces and displacements for H2 Optimization
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Jmol and File Link
H2 Optimisation |
H2 Vibration Frequencies
| Mode # | Frequency | Infrared |
|---|---|---|
| 1 | 4465.68 | 0.0000 |
Haber-Bosch reaction energy calculations
E(NH3) = -56.55776873 a.u.
2*E(NH3) = -113.11553746 a.u.
E(N2) = -109.52412868 a.u.
E(H2) = -1.17853936 a.u.
3*E(H2) = -3.53561808 a.u.
ΔE=2*E(NH3) - [E(N2) + 3*E(H2)] = -0.0557907 a.u. = -146.47849401 kJ mol-1
O2 molecule
Optimized O-O bond distance = 1.21602 Å
Optimized O-O bond angle = 180.000 °
Calculation method = RB3LYP
Basis set = 6-31G(d.p)
Final energy E(RB3LYP) = -150.25742434 a.u.
RMS gradient norm = 0.00007502 a.u.
Point group = D∞H
Final set of forces and displacements for O2 Optimization
Item Value Threshold Converged?
Maximum Force 0.000130 0.000450 YES
RMS Force 0.000130 0.000300 YES
Maximum Displacement 0.000080 0.001800 YES
RMS Displacement 0.000113 0.001200 YES
Jmol and File Link
O2 Optimisation |
O2 Vibration Frequencies
| Mode # | Frequency | Infrared |
|---|---|---|
| 1 | 1642.74 | 0.0000 |
MO's of Optimized O2
Energy = -1.27663 a.u.
Is the MO occupied or unoccupied? = Occupied.
Is the MO bonding or antibonding? = Bonding 2σg
What AO's contribute to the MO? = Two 2s AO's, one from each oxygen atom.
What effect does the MO have on bonding? = This MO has no contribution on the bonding between the two oxygen atoms.
Energy = -0.53151 a.u.
Is the MO occupied or unoccupied? = Occupied.
Is the MO bonding or antibonding? = Bonding 3σg
What AO's contribute to the MO? = Two 2pz AO's, one from each oxygen atom.
What effect does the MO have on bonding? = This MO has no contribution on the bonding between the two oxygen atoms.
Energy = -0.51526 a.u.
Is the MO occupied or unoccupied? = Occupied.
Is the MO bonding or antibonding? = Bonding 1πu
What AO's contribute to the MO? = Two 2px AO's or two 2py AO's, one from each oxygen atom.
What effect does the MO have on bonding? = This MO forms the sigma bond between the two oxygen atoms.
Energy = -0.25018 a.u.
Is the MO occupied or unoccupied? = Occupied.
Is the MO bonding or antibonding? = Antibonding 1π*g
What AO's contribute to the MO? = Two 2px AO's or two 2py AO's, one from each oxygen atom.
What effect does the MO have on bonding? = This MO has no contribution on the bonding between the two oxygen atoms.
Gauss View shows this MO as being filled with 2 electrons of opposite spin, therefore identifying it as the HOMO. However the 1π*g is made of two degenerate energy levels, therefore the two electrons are actually unpaired, making this MO the SOMO. As a result of this, oxygen is para magnetic.
Energy = -0.17928 a.u.
Is the MO occupied or unoccupied? = Unoccupied.
Is the MO bonding or antibonding? = Anti bonding 1π*g, this MO is the LUMO
What AO's contribute to the MO? = Two 2px AO's or two 2py AO's, one from each oxygen atom.
What effect does the MO have on bonding? = This MO has no contribution on the bonding between the two oxygen atoms.
CF4 molecule
Optimized C-F bond distance = 1.32939 Å
Optimized F-C-F bond angle = 109.471 °
Calculation method = RB3LYP
Basis set = 6-31G(d.p)
Final energy E(RB3LYP) = -437.47627267 a.u.
RMS gradient norm = 0.00004049 a.u.
Point group = TD
Final set of forces and displacements for CF4 Optimization
Item Value Threshold Converged?
Maximum Force 0.000078 0.000450 YES
RMS Force 0.000042 0.000300 YES
Maximum Displacement 0.000133 0.001800 YES
RMS Displacement 0.000071 0.001200 YES
Jmol and File Link
CF4 Optimisation |
CF4 Vibration Frequencies
| Mode # | Frequency | Infrared |
|---|---|---|
| 1 | 424.00 | 0.0000 |
| 2 | 424.00 | 0.0000 |
| 3 | 616.80 | 4.5048 |
| 4 | 616.80 | 4.5048 |
| 5 | 616.80 | 4.5048 |
| 6 | 906.23 | 0.0000 |
| 7 | 1305.80 | 387.1737 |
| 8 | 1305.80 | 387.1737 |
| 9 | 1305.80 | 387.1737 |
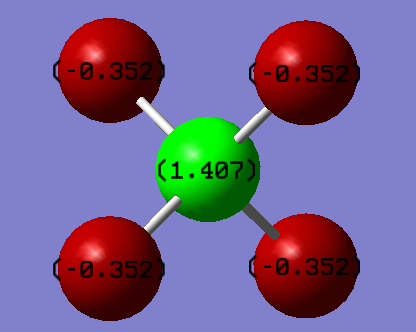
Charge Distribution of CF4
The fluorine's are more electronegative than the carbon, therefore will withdraw electron density from the carbon atom. As a result of this, the fluorine's will be negatively charged and the carbon will be positive, which is confirmed by the charge distribution shown above.