Rep:Mod:michellesimon
NH3
Calculation method: RB3LYP
Basis set: 6-31G(d,p)
E(RB3LYP) -56.55776873 a.u.
RMS Gradient Norm 0.00000323 a.u.
Point Group: C3V
Optimised angle: 105.7
Optimised length: 1.018 Angstroms
Item Value Threshold Converged?
Maximum Force 0.000006 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000014 0.001800 YES
RMS Displacement 0.000009 0.001200 YES
Predicted change in Energy=-1.141618D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7446 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7446 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7446 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8637 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
NH3 |
Gaussian log file can be accessed here: Media:MICHELLESIMON_NH3_OPT_FPOP.LOG
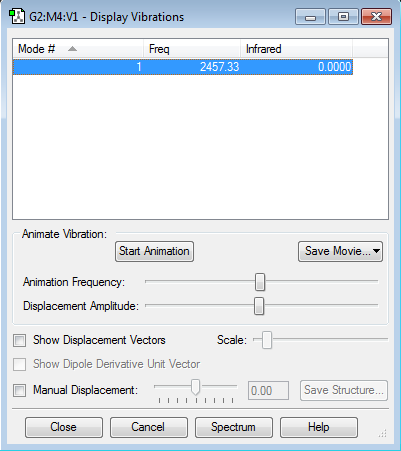
Vibrations
How many modes do you expect from the 3N-6 rule? 6
Which modes are degenerate (ie have the same energy)? 2&3 and 5&6
Which modes are "bending" vibrations and which are "bond stretch" vibrations?
Bending: 1, 2, 3
Bond stretch: 4, 5, 6
Which mode is highly symmetric? 4
One mode is known as the "umbrella" mode, which one is this? 1
How many bands would you expect to see in an experimental spectrum of gaseous ammonia? 2
Charge distribution
H: 0.375
N: -1.125
N is expected to have a slightly negative charge because it has more protons in its nucleus and should draw bonding electrons closer to itself, and H should have a slightly positive charge as it it poorer at attracting bonding pairs. ie. N is more electronegative than H
N2
Calculation Method RB3LYP
Basis Set 6-31G(d,p)
E(RB3LYP) -109.52359111 a.u.
RMS Gradient Norm 0.02473091 a.u.
Point group D∞h
Bond length 1.10 Angstroms
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.401047D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1055 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
N2 |
Gaussian log file can be accessed here: Media:MS4516_N2_OPT.LOG
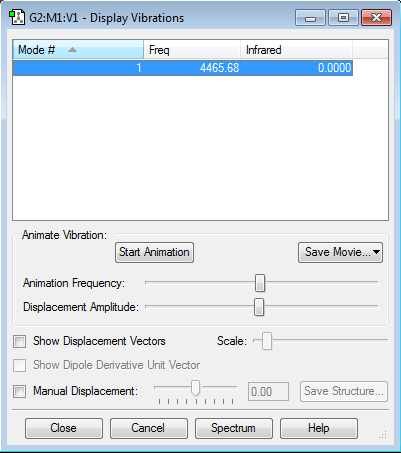
Vibrations
H2
Calculation Method RB3LYP
Basis Set 6-31G(d,p)
E(RB3LYP) -1.17853936 a.u.
RMS Gradient Norm 0.00000017 a.u.
Point group D∞h
Bond length 0.743 Angstroms
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 0.7428 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
H2 |
Gaussian log file can be accessed here: Media:MS4516 H2 OPT.LOG
Vibrations
Reactivity
E(NH3)= -56.55776873 a.u.
2*E(NH3)= -113.11553746 a.u.
E(N2)= -109.52359111 a.u.
E(H2)= -1.17853936 a.u.
3*E(H2)= -3.53561808 a.u.
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.05632827 a.u.
Which is -147.89 kJ/mol. (This is for 2 molecules of NH3)
The ammonia product is more stable because the reaction is exothermic (negative energy change), which means that in converting N2 and H2 into NH3, excess energy was released from the system into the surroundings. Therefore ammonia is at a lower energy than the reactants.
Molar ΔE = -73.945 kJ/mol
This molar ΔE compared to its literature value is higher (-41kJ/mol) [1] . This is likely to be due to the inaccuracies in calculations of the energy. Experimentally there are many more small effects on the energy of the molecule that have not been considered in RB3LYP calculations, especially since the processing power of the computer used is quite low. Therefore this causes a discrepancies in the two values.
- ↑ T. Lister, J. Renshaw, in New Understanding Chemistry for Advanced Level, Nelson Thornes, 2000, ch. 11, pp. 137
Own molecule 1: BH4-
Calculation Method RB3LYP
Basis Set 6-31G(d,p)
Charge -1
E(RB3LYP) -27.24992701 a.u.
RMS Gradient Norm 0.00000589 a.u.
B-H bond length 1.23940A
H-B-H bond angle 109.47
Point group Td
Item Value Threshold Converged?
Maximum Force 0.000011 0.000450 YES
RMS Force 0.000006 0.000300 YES
Maximum Displacement 0.000057 0.001800 YES
RMS Displacement 0.000030 0.001200 YES
Predicted change in Energy=-1.299237D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.2394 -DE/DX = 0.0 !
! R2 R(1,3) 1.2394 -DE/DX = 0.0 !
! R3 R(1,4) 1.2394 -DE/DX = 0.0 !
! R4 R(1,5) 1.2394 -DE/DX = 0.0 !
! A1 A(2,1,3) 109.4712 -DE/DX = 0.0 !
! A2 A(2,1,4) 109.4712 -DE/DX = 0.0 !
! A3 A(2,1,5) 109.4712 -DE/DX = 0.0 !
! A4 A(3,1,4) 109.4712 -DE/DX = 0.0 !
! A5 A(3,1,5) 109.4712 -DE/DX = 0.0 !
! A6 A(4,1,5) 109.4712 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -120.0 -DE/DX = 0.0 !
! D2 D(2,1,5,3) 120.0 -DE/DX = 0.0 !
! D3 D(2,1,5,4) -120.0 -DE/DX = 0.0 !
! D4 D(3,1,5,4) 120.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
BH4- |
Gaussian log file can be accessed here: Media:MS4516_BH4 OPT.LOG
Charge distribution
H: -0.096
B: -0.615
B and H have very similar electronegativities, therefore charges on the atoms are fairly similar.
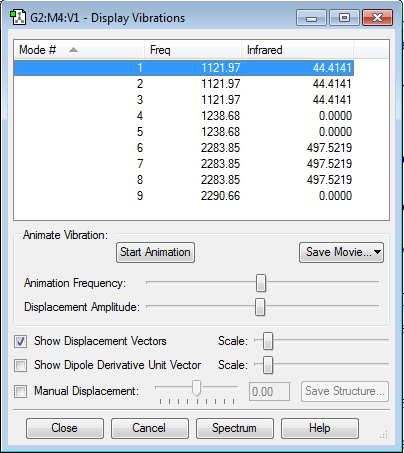
Vibrations
-Two peaks should be seen on the infrared spectrum for BH4-.
- One at 1122.0cm-1 and another at 2283.9cm-1
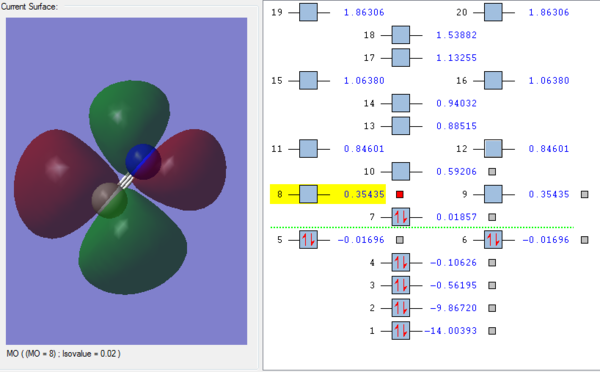
MOs
Own molecule 2: CN-
Calculation Method RB3LYP
Basis Set 6-31G(d,p)
Charge -1
E(RB3LYP) -92.82453153 a.u.
RMS Gradient Norm 0.00000704 a.u.
C-N bond length 1.18409A
Point group C∞v
Item Value Threshold Converged?
Maximum Force 0.000012 0.000450 YES
RMS Force 0.000012 0.000300 YES
Maximum Displacement 0.000005 0.001800 YES
RMS Displacement 0.000008 0.001200 YES
Predicted change in Energy=-6.650389D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1841 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
CN- |
Gaussian log file can be accessed here: Media:MS4516_CN OPT.LOG
Charge distribution
C: -0.246
N: -0.754
- As with NH3, the nitrogen atom is more electronegative than carbon is.
- Therefore it pulls electrons closer to its nucleus than carbon does, resulting in a more negative charge distributed onto the N atom.
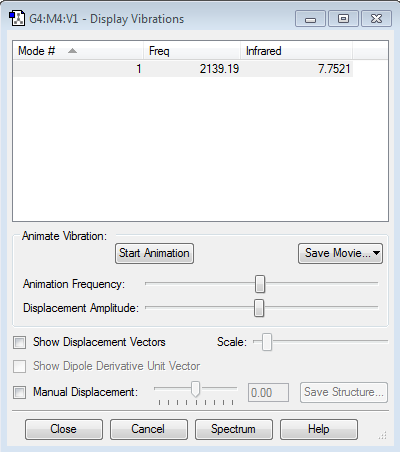
Vibrations
- Although CN- is also a linear molecule (like H2 and N2 previously explored), there is one peak observed on the infrared spectrum for CN- at 2139.1cm-1.
- This is due to a change in dipole moment during the vibration, which differs from H2 and N2 homodiatomic molecules because CN- is composed of two different atoms (heterodinuclear).