Rep:Mod:mhl12comp
Comparison of BH3, BBr3 and GaBr3
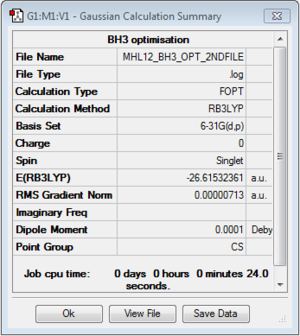
BH3
B3LYP/3-21G level
Optimisation log file here
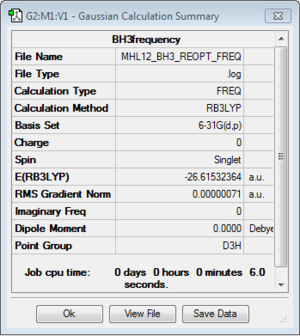
B3LYP/6-31G(d,p) level
Optimisation log file here
| summary data | convergence | Jmol | |||
|---|---|---|---|---|---|
|
Item Value Threshold Converged? Maximum Force 0.000203 0.000450 YES RMS Force 0.000098 0.000300 YES Maximum Displacement 0.000867 0.001800 YES RMS Displacement 0.000415 0.001200 YES |
|
Geometry Data
| r(E-X) Å | 1.19 |
| θ(X-E-X) degrees(º) | 120.0 |
Frequency Analysis
Frequency file: here
| summary data | low modes |
|---|---|
|
Low frequencies --- -10.1940 -10.1821 -3.1776 -0.0012 0.0579 0.4920 Low frequencies --- 1162.9850 1213.1461 1213.1463 |
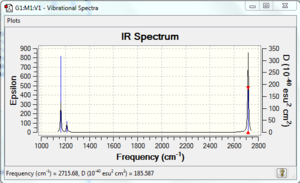
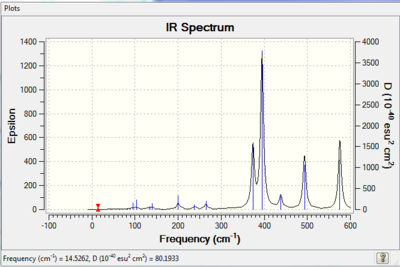
Vibrational spectrum for BH3
| wavenumber | Intensity | IR active? | type |
| 1163 | 93 | yes | bend |
| 1213 | 14 | no | bend |
| 1213 | 14 | very slight | bend |
| 2583 | 0 | no | stretch |
| 2716 | 126 | yes | stretch |
| 2716 | 126 | yes | stretch |
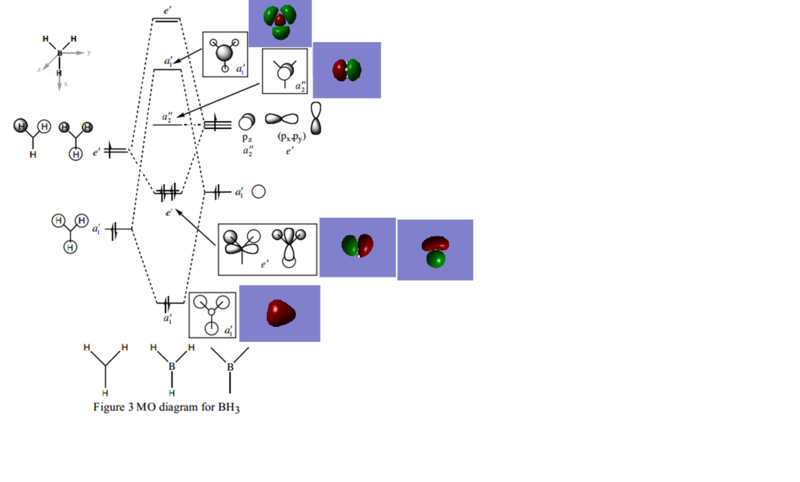
Calculated MO and LCAO
MO checkpoint file: file
LCAO MO (Credit to Dr Tricia Hunt, diagram adapted from Year 2 Lecture 4 Tutorial Problem Model Answers)
The calculated MO agrees with the LCAO approximation.
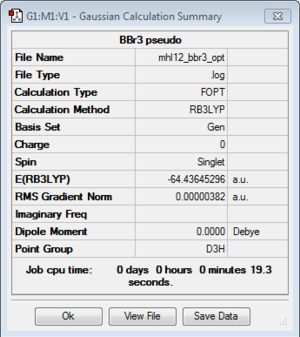
BBr3
B3LYP/6-31(d,p)LANL2DZ
Optimisation log file here
| summary data | convergence | Jmol | |||
|---|---|---|---|---|---|
|
Item Value Threshold Converged? Maximum Force 0.000008 0.000450 YES RMS Force 0.000005 0.000300 YES Maximum Displacement 0.000036 0.001800 YES RMS Displacement 0.000023 0.001200 YES |
|
Geometry Data
| r(E-X) Å | 1.93 |
| θ(X-E-X) degrees(º) | 120.0 |
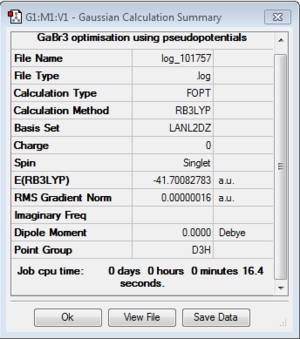
GaBr3
B3LYP/LanL2DZ
Optimisation log file here
| summary data | convergence | Jmol | |||
|---|---|---|---|---|---|
|
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000003 0.001800 YES RMS Displacement 0.000002 0.001200 YES |
|
Geometry Data
| r(E-X) Å | 2.35 |
| θ(X-E-X) degrees(º) | 120.0 |
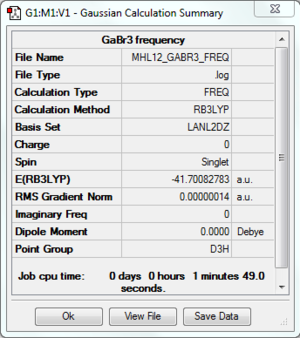
Frequency Analysis
Frequency file: here
| summary data | low modes |
|---|---|
|
Low frequencies --- -0.5293 -0.5288 -0.0024 -0.0010 0.0233 1.1993 Low frequencies --- 76.3744 76.3753 99.6981 |
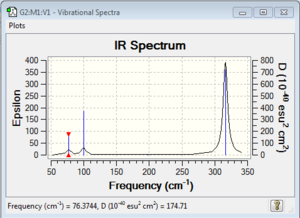
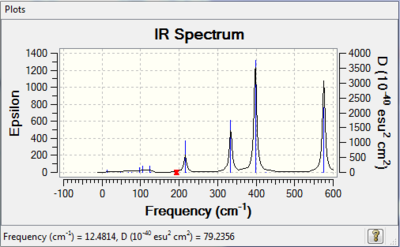
Vibrational spectrum for GaBr3
| wavenumber | Intensity | IR active? | type |
| 76 | 3 | no | bend |
| 76 | 3 | no | bend |
| 100 | 9 | very slight | bend |
| 197 | 0 | no | stretch |
| 316 | 57 | yes | stretch |
| 316 | 57 | yes | stretch |
Structural Comparison
Geometry Data
| BH3 | BBr3 | GaBr3 | |
|---|---|---|---|
| r(E-X) Å | 1.19 | 1.93 | 2.35 |
| θ(X-E-X) degrees(º) | 120.0 | 120.0 | 120.0 |
Discussion:
In GaussView, when it thinks the bond length is too long (than its threshold) or the bonding energy is unfavorable, it will not show the bond. This always happens during the optimization process, when GaussView try to optimize the bond length.
We can see that the bond length increases from B-H (BH3) to B-Br (BBr3) This is mostly due to the increase in radii of ligands; the mismatch in orbital size causes weaker interaction and thus a larger bond length. Boron and Gallium both belong to Group 13. Here again, down the group radii of Ga is larger than that of B. Hence the bond length of Ga-Br is larger than that of B-Br. However comparing the increment of B-H to B-Br (0.74Å) and B-Br to Ga-Br(0.42Å), the effect of changing a different nature or substantially larger ligand lead to a larger change in bond distance. Chaging the centre atom will lead to corresponding change according to their ionic radii.
Definition of chemical bond (from [[1]]) : When forces acting between two atoms or groups of atoms lead to the formation of a stable independent molecular entity, a chemical bond is considered to exist between these atoms or groups. The principal characteristic of a bond in a molecule is the existence of a region between the nuclei of constant potential contours that allows the potential energy to improve substantially by atomic contraction at the expense of only a small increase in kinetic energy. Not only directed covalent bonds characteristic of organic compounds, but also bonds such as those existing between sodium cations and chloride anions in a crystal of sodium chloride or the bonds binding aluminium to six molecules of water in its environment, and even weak bonds that link two molecules of O2 into O4, are to be attributed to chemical bonds. [1] In simple, chemical bond is a 'link' between two atoms/groups, with attractive forces acting on both atoms/groups.
For covalent bond: Taking silica, a stable and abundant compound on Earth, as a benchmark, Si-O having a bond energy of 452 kJ/mol; so bonding energy above 450 kJ/mol, such as multiple bonding C≡N, N≡N, C=O etc., could be considered as very strong bond; however for single bonding, Si-O is one of the strongest bond. Typical C-C bond has a value of 346 kJ/mol which is around 20% lower in energy than that of Si-O. C-C still could be considered a strong bond. Bonding energies which is 50% less than Si-O (say 200 kJ/mol) are considered medium bond. N-N, N-B etc. have bonding energies of 100+ kJ/mol. Bonding energies which is below 100 kJ/mol (an order of magnitude lower than majority of covalent bonding energy)
For ionic bond: Ionic bonds generally have higher bonding energy, ranging from 700-4000 [2]
For dative bond: Dative bond is a slightly special case. It is a covalent bond but with a significant dipole on the chemical bond. The more electronegative atom will carry partial negative charge and the more electropositive atom will carry partial positive charge.
However, usually molecules have the characteristics of both ionic bond and covalent bond in itself. Hence theoretical calculation of bond energy does not always agree with the observed bond energy.
Vibrational Comparison
Discussion: Anharmonic oscillator/ Morse curve, a better empirical curve fits the experimental better, but in our frequency analysis, we assumed that the vibration works under harmonic oscillation;this means that if we stretch/compress the spring and release it, it will oscillate around its equilibrium position sinusoidally.
F=-ks(r-r0), where ks is the spring constant, r is the length of extension/compression, r0 is the equilibrium length.
ω=(ks/μ)0.5, where μ=reduced mass
ευ=[(υ+1/2)ω]/(2πc) where υ= 0,1,2,3,......
Bond length of B-H is considerably shorter than that of Ga-Br. Generally, increment in bond length reduces the bond strength, or in this case we can treat bond strength as 'spring constant', and hence reduces the vibrational frequency. Of course, the reduced mass of both systems does affect the vibrational frequency as well. The higher the reduced mass, Ga-Br(36.8) compare to B-H(0.9), the lower the frequency of wave number. This is rationalized using Hooke's law without considering any relativistic effect or isotopes effect. It agrees with our calculation; GaBr3 IR spectrum has bands ranging from 70-300, while BH3 IR spectrum ranges from 1000-3000
The frequency of A2" umbrella motion of BH3 and GaBr3 1163 and 100 respectively. The difference in frequencies has been explained above. Umbrella motion of BH3 is mode #1 with a high intensity (93), while umbrealla motion of GaBr3 is mode #3 with low intensity (9). This is most probably due to a change in reduced mass and bond strength; leading to reorder of vibration modes. The intensity of the Ir spectrum relates to the amplitude of oscillation. The higher the amplitude the higher larger the intensity. And comparing the vibrating animation of both molecules, in BH3, Boron and hydrogen did vibrate relative to each other, while in the case of GaBr3 only Ga is moving. Usually, the stom with relatively lighter mass will vibrate will a larger amplitude than the one that is heavier.
In this lab, all comparison should be run under same basis set. This is to compare two things in a same manner; basis could be thought as a fixed variable, and an experiment should only have one responding variable. Frequency analysis is to make sure the optimisation process does provide a minima. Low frequencies in the calculation represents translational and rotational motion which should be excluded from the vibrational spectrum.
Bonding analysis of NH3BH3
BH3
B3LYP/6-31G(d,p) level optimisation and frequency analysis, refer to here
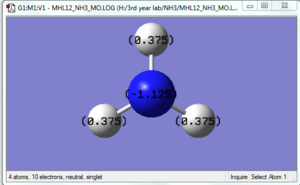
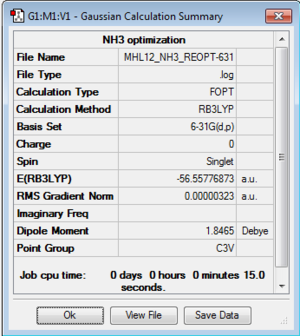
NH3
B3LYP/6-31G(d,p) level
Optimisation log file here
| summary data | convergence | Jmol | |||
|---|---|---|---|---|---|
|
Item Value Threshold Converged? Maximum Force 0.000006 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000012 0.001800 YES RMS Displacement 0.000008 0.001200 YES |
|
Geometry Data
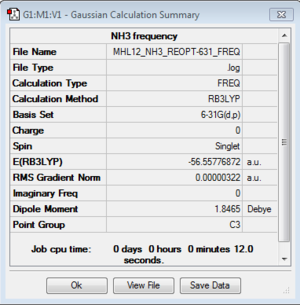
Frequency
Frequency file: here
| summary data | low modes |
|---|---|
|
Low frequencies --- -0.0129 -0.0027 0.0005 7.0724 8.1020 8.1023 Low frequencies --- 1089.3849 1693.9369 1693.9369 |
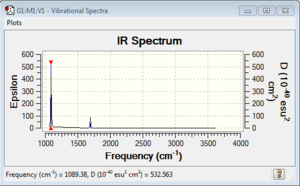
Vibrational spectrum for NH3
| wavenumber | Intensity | IR active? | type |
| 1089 | 145 | yes | bend |
| 1694 | 14 | very slight | bend |
| 1694 | 14 | very slight | bend |
| 3461 | 1 | no | stretch |
| 3590 | 0 | no | stretch |
| 3590 | 0 | no | stretch |
NH3 with C3V point group has relatively small number of IR active bands.
Population Analysis
NBO analysis log: here
Nitrogen is a more electronegative atoms and hence has a more negative dipole. And since NH3 is a neutral molecule, hence the dipole will adds up and give 0.
Bond orbital/ Coefficients/ Hybrids
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.99909) BD ( 1) N 1 - H 2
( 68.83%) 0.8297* N 1 s( 24.86%)p 3.02( 75.05%)d 0.00( 0.09%)
0.0001 0.4986 0.0059 0.0000 0.0000
0.0000 0.8155 0.0277 -0.2910 0.0052
0.0000 0.0000 -0.0281 -0.0087 0.0013
( 31.17%) 0.5583* H 2 s( 99.91%)p 0.00( 0.09%)
0.9996 0.0000 0.0000 -0.0289 0.0072
2. (1.99909) BD ( 1) N 1 - H 3
( 68.83%) 0.8297* N 1 s( 24.86%)p 3.02( 75.05%)d 0.00( 0.09%)
0.0001 0.4986 0.0059 0.0000 -0.7062
-0.0239 -0.4077 -0.0138 -0.2910 0.0052
0.0076 0.0243 0.0140 0.0044 0.0013
( 31.17%) 0.5583* H 3 s( 99.91%)p 0.00( 0.09%)
0.9996 0.0000 0.0250 0.0145 0.0072
3. (1.99909) BD ( 1) N 1 - H 4
( 68.83%) 0.8297* N 1 s( 24.86%)p 3.02( 75.05%)d 0.00( 0.09%)
0.0001 0.4986 0.0059 0.0000 0.7062
0.0239 -0.4077 -0.0138 -0.2910 0.0052
-0.0076 -0.0243 0.0140 0.0044 0.0013
( 31.17%) 0.5583* H 4 s( 99.91%)p 0.00( 0.09%)
0.9996 0.0000 -0.0250 0.0145 0.0072
4. (1.99982) CR ( 1) N 1 s(100.00%)
1.0000 -0.0002 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
5. (1.99721) LP ( 1) N 1 s( 25.38%)p 2.94( 74.52%)d 0.00( 0.10%)
0.0001 0.5036 -0.0120 0.0000 0.0000
0.0000 0.0000 0.0000 0.8618 -0.0505
0.0000 0.0000 0.0000 0.0000 -0.0310
6. (0.00000) RY*( 1) N 1 s( 99.98%)p 0.00( 0.02%)d 0.00( 0.00%)
BD refers to bonding orbital, CR refers to core orbital and LP refers to lone pair. The first, second and third BD shows that N-H orbital, which 69% contributed by nitrogen atoms. And nitrogen orbital is sp3 hybridized while H orbital is purely s. The core orbital of nitrogen are purely s. The lone pair of nitrogen is sp3 hybridised as well.
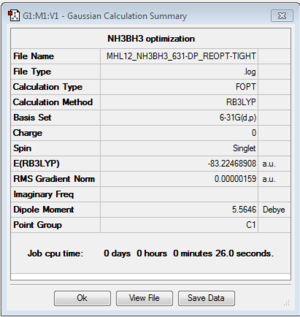
NH3BH3
B3LYP/6-31G(d,p) level
Optimisation log file here
| summary data | convergence | Jmol | |||
|---|---|---|---|---|---|
|
Item Value Threshold Converged? Maximum Force 0.000003 0.000015 YES RMS Force 0.000001 0.000010 YES Maximum Displacement 0.000022 0.000060 YES RMS Displacement 0.000010 0.000040 YES |
|
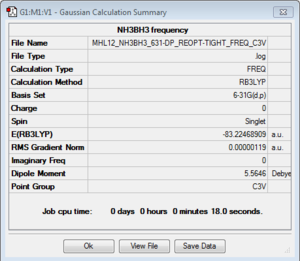
Frequency analysis
Frequency file: here
| summary data | low modes |
|---|---|
|
Low frequencies --- -5.8792 -0.3571 -0.0537 -0.0014 1.0077 1.1094 Low frequencies --- 263.2802 632.9504 638.4510 |
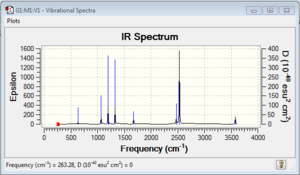
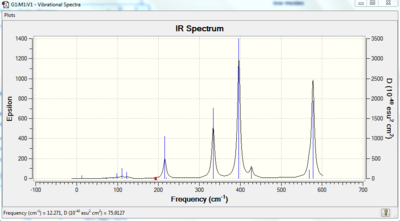
Vibrational spectrum for NH3BH3
| wavenumber | Intensity | IR active? | type |
| 262 | 0 | no | bend |
| 633 | 14 | very slight | stretch |
| 638 | 4 | no | bend |
| 638 | 4 | no | bend |
| 1069 | 41 | yes | bend |
| 1069 | 41 | yes | bend |
| 1069 | 41 | yes | bend |
| 1196 | 109 | yes | bend |
| 1204 | 3 | no | bend |
| 1204 | 3 | no | bend |
| 1676 | 28 | yes | bend |
| 1676 | 28 | yes | bend |
| 2472 | 67 | yes | stretch |
| 2532 | 231 | yes | stretch |
| 2532 | 231 | yes | stretch |
| 3464 | 3 | no | stretch |
| 3581 | 28 | yes | stretch |
| 3581 | 28 | yes | stretch |
Bond Energy
E(NH3)= -56.55776873 a.u.
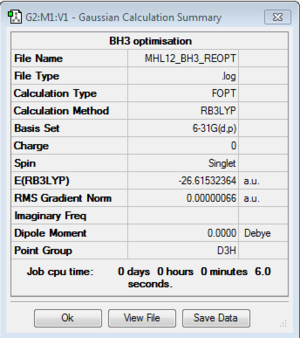
E(BH3)= -26.61532364 a.u.
E(NH3BH3)= -83.22468908 a.u.
ΔE=E(NH3BH3) - [E(NH3)+E(BH3)]
= -83.22468908 a.u. - [-56.55776873 a.u. -26.61532364 a.u.] = -0.05159671 a.u. = -135.47 kJ/mol (2 d.p.)
Discussion: As this N->B a dative covalent bond, we are comparing N-B bond with normal type of covalent bond. C-C bond which is isoelectronic to N-B bond; typical C-C bond has a bond energy of 346kJ/mol. Hence N-B bond is slightly weaker bond relative to C-C bond. Considering that bond energy of N-B is still of the same order of magnitude with C-C bond, and the fact that the bond energy is way above interaction of hydrogen bonding (typical value 8-30 kJ/mol, except F-H-F), it can be said that N-B bond is a medium(strength) bonding.
2nd Week Project - Lewis Acids and Bases
Optimization of Al2Cl2Br4
The optimization process was computed using 'GEN' basis set and keywords pseudo=cards gfinput. This allow us to specify basis set for each atoms individually. The following instructions were added into the gjf files:
Al 0 6-31G(d,p) **** Cl 0 6-31G(d,p) **** Br 0 LanL2DZ **** Br 0 LanL2DZ
| Compound | Isomer 1 | Isomer 2 | Isomer 3 | Isomer 4 | ||||||||
|
|
|
| ||||||||||
| Covergence Data |
Item Value Threshold Converged? Maximum Force 0.000005 0.000015 YES RMS Force 0.000001 0.000010 YES Maximum Displacement 0.000008 0.000060 YES RMS Displacement 0.000003 0.000040 YES |
Item Value Threshold Converged? Maximum Force 0.000002 0.000015 YES RMS Force 0.000001 0.000010 YES Maximum Displacement 0.000017 0.000060 YES RMS Displacement 0.000007 0.000040 YES |
Item Value Threshold Converged? Maximum Force 0.000001 0.000015 YES RMS Force 0.000000 0.000010 YES Maximum Displacement 0.000018 0.000060 YES RMS Displacement 0.000007 0.000040 YES |
Item Value Threshold Converged? Maximum Force 0.000001 0.000015 YES RMS Force 0.000000 0.000010 YES Maximum Displacement 0.000016 0.000060 YES RMS Displacement 0.000005 0.000040 YES | ||||||||
| File Type | .log | .log | .log | .log | ||||||||
| Calculation Type | FOPT | FOPT | FOPT | FOPT | ||||||||
| Calculation Method | RB3LYP | RB3LYP | RB3LYP | RB3LYP | ||||||||
| Basis Set | Gen | Gen | Gen | Gen | ||||||||
| Charge | 0 | 0 | 0 | 0 | ||||||||
| Spin | Singlet | Singlet | Singlet | Singlet | ||||||||
| E(RB3LYP) / a.u. | -1458.35116016 | -1458.34598708 | -1458.34122428 | -1458.34120346 | ||||||||
| RMS Gradient Norm / a.u. | 0.00000222 | 0.00000255 | 0.00000034 | 0.00000034 | ||||||||
| Imaginary Freq | ||||||||||||
| Dipole Moment / Debye | 0.0002 | 0.1240 | 0.0002 | 0.1707 | ||||||||
| Point Group | C1 | C1 | C1 | C1 | ||||||||
| Job cpu time | 0 days 0 hours 5 minutes 38.8 seconds | 0 days 0 hours 17 minutes 56.6 seconds | 0 days 0 hours 10 minutes 9.1 seconds | 0 days 0 hours 12 minutes 48.1 seconds | ||||||||
| D-Space Link | DOI:10042/107512 | DOI:10042/107510 | DOI:10042/107508 | DOI:10042/107506 |
All calculations have been confirmed with the convergence of the data. The optimization will then be further confirmed with frequency analysis (±15cm-1 for the low frequencies mode as a benchmark to check the optimization process)
| Computational bond length / Å | |
| Terminal Br-Al | 2.28 |
| Terminal Cl-Al | 2.09 |
| Bridging Br-Al | 2.49 |
| Bridging Cl-Al | 2.30 |
Bridging atom and aluminium has a longer bond length that of terminal Al-halides bond indicates that it is not a conventional 2c-2e bond.
Discussion
| Energy reported / a.u. | Relative Energy / a.u. | Relative Energy / KJ/mol | |
| Isomer 1 | -1458.35116016 | 0 | 0 |
| Isomer 2 | -1458.34598708 | 0.00517308 | 13.58 |
| Isomer 3 | -1458.34122428 | 0.00993588 | 26.14 |
| Isomer 4 | -1458.34120346 | 0.00995670 | 26.14 |
Isomer has the lowest energy indicates that it is the most stable isomer. As we exchange one bridging Cl with one terminal Br, the energy increases by 13.58 kJ/mol, and increases another 13.06 kJ/mol to 26.14 kJ/mol as we remove one more bridging Cl. And the cis/trans configuration of Cl (isomer 3 and 4) does not have significant changes in energy. From the evidence above, it is likely that Cl is more favourable as bridging atom than Br. This is probably due to Cl, with higher electronegativities, are more compatible with the high lewis acidity Al atom.
Dissociation energy
E(Al2Cl2Br4)= -1458.35116016 a.u.
E(AlClBr2)= -26.61532364 a.u. *Refer to DOI:10042/107530
ΔE=E(Al2Cl2Br4) - 2*E(AlClBr2)
= -1458.35116016 a.u. - 2*[ -729.15823878 a.u.] = -0.0346826 a.u. = -91.06 kJ/mol (2 d.p.)
Frequency Analysis
| Compound | Isomer 1 | Isomer 2 | Isomer 3 | Isomer 4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Calculation Method | RB3LYP | RB3LYP | RB3LYP | RB3LYP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Basis Set | Gen | Gen | Gen | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge | 0 | 0 | 0 | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spin | Singlet | Singlet | Singlet | Singlet | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| E(RB3LYP) / a.u. | -1458.35116016 | -1458.34598708 | -1458.34122763 | -1458.34120346 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| RMS Gradient Norm / a.u. | 0.0000441 | 0.00000255 | 0.00000753 | 0.00000064 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Imaginary Freq | 0 | 0 | 0 | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dipole Moment / Debye | 0.0000 | 0.1240 | 0.0000 | 0.1707 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Point Group | D2H | C1 | C2H | C2V | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Job cpu time | 0 days 0 hours 0 minutes 50.0 seconds | 0 days 0 hours 2 minutes 22.4 seconds | 0 days 0 hours 1 minutes 0.9 seconds | 0 days 0 hours 1 minutes 1.7 seconds. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Low Modes |
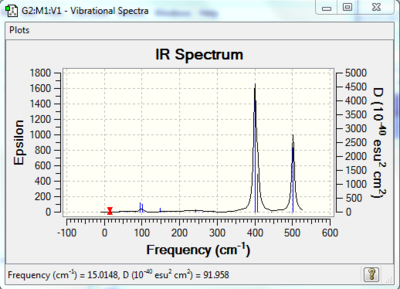
Low frequencies --- -3.5251 -2.2048 -0.0030 -0.0022 -0.0019 1.9779 Low frequencies --- 15.0148 38.6160 59.6441 |
Low frequencies --- -2.6621 -0.0021 -0.0018 0.0018 2.1611 3.3401 Low frequencies --- 14.5344 44.1267 64.0506 |
Low frequencies --- -5.0200 -3.3842 -3.0570 0.0035 0.0039 0.0041 Low frequencies --- 12.4814 47.7011 67.3961 |
Low frequencies --- -4.8432 -3.3720 -3.0141 -0.0025 -0.0016 0.0013 Low frequencies --- 12.2710 49.9139 71.1649 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| D-Space Link | DOI:10042/107492 | DOI:10042/107490 | DOI:10042/107488 | DOI:10042/107486 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IR Spectra |
|
|
|
|
Discussion
A molecule must have stretch or bent and created a change in dipole in order to be IR active. Hence, the higher the symmetry of molecules, the less IR active band exist due to many symmetric stretches and bent will not create a change in dipole.
The point groups of the four isomers above are highlighted in the table. Isomer 2 belongs to C1 group, the group with only E symmetry elements, thus it has the most IR bands. The other 3 isomers have higher symmetry, hence they have less IR bands.
Bridging halide- Al stretch gives a lower wavenumber, it is most probably due to the reasoning that the bond length has become substantially longer than that of terminal halide-Al. The reasoning could be found on the discussion of vibrational frequency of Ga-Br and B-H. And chlorine vibrate more strongly than bromine, most probably due to their masses.
In the IR spectrum from above, for example in spectrum in isomer 1, for the same symmetrical stretching of Cl- Al, it can be due to vibrational movement of Cl or Al or both. They have different frequencies and intensities. The reordering of vibrational modes make it hard to analyse and we have to examine all vibrational movement one by one. Bridging halides vibrate in a smaller amplitude than terminal halide; this is most probably due to the 3c-4e bonding.
The cis trans configuration does not affect the IR spectrum significantly. They both have similar stretches at similar frequencies with similar intensities. The reorder of modes does not manifest in this case is due to the fact that both the molecules have the same element in it. And the configuration of the molecules is very similar in a way that the bridging molecules remain to be Br, only changing the position of terminal halides.
MO Analysis
5 MOs of Isomer 1 of Al2Cl2Br4 are investigated for this project. All files for MO analysis can be obtained from DOI:10042/107495
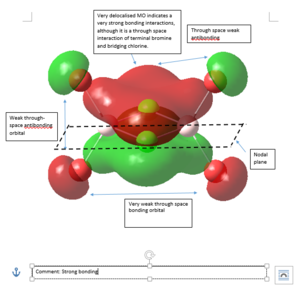
|
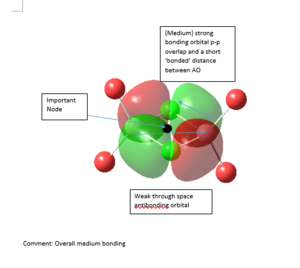
|

|
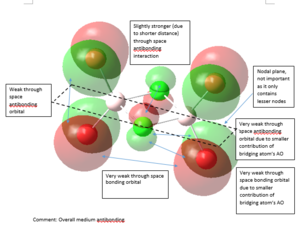
|
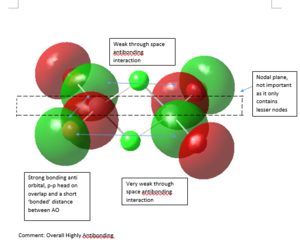
|
Strong bonding -> Medium bonding -> Weak bonding -> Medium antibonding -> Strong antibonding
NBO analysis
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.96691) BD ( 1)Al 1 -Cl 3
( 12.14%) 0.3484*Al 1 s( 20.28%)p 3.77( 76.46%)d 0.16( 3.26%)
0.0000 -0.0011 0.4501 0.0142 0.0003
-0.5362 -0.0191 0.0000 0.0000 0.0000
0.0002 0.6887 0.0496 0.0000 -0.1524
0.0000 0.0816 0.0519
( 87.86%) 0.9374*Cl 3 s( 23.62%)p 3.23( 76.20%)d 0.01( 0.18%)
0.0000 -0.0002 0.4859 0.0120 0.0000
0.7062 0.0012 0.0000 0.0000 0.0000
0.0000 -0.5131 0.0036 0.0000 -0.0356
0.0000 0.0121 0.0191
2. (1.96691) BD ( 1)Al 1 -Cl 4
( 12.14%) 0.3484*Al 1 s( 20.28%)p 3.77( 76.46%)d 0.16( 3.26%)
0.0000 -0.0011 0.4501 0.0142 0.0003
-0.5362 -0.0191 0.0000 0.0000 0.0000
-0.0002 -0.6887 -0.0496 0.0000 0.1524
0.0000 0.0816 0.0519
( 87.86%) 0.9374*Cl 4 s( 23.62%)p 3.23( 76.20%)d 0.01( 0.18%)
0.0000 -0.0002 0.4859 0.0120 0.0000
0.7062 0.0012 0.0000 0.0000 0.0000
0.0000 0.5131 -0.0036 0.0000 0.0356
0.0000 0.0121 0.0191
3. (1.96807) BD ( 1)Al 1 -Br 7
( 21.17%) 0.4601*Al 1 s( 29.66%)p 2.28( 67.62%)d 0.09( 2.73%)
0.0000 -0.0001 0.5445 -0.0056 0.0001
0.4361 0.0324 0.0000 -0.6941 -0.0568
0.0000 0.0000 0.0000 -0.1228 0.0000
0.0000 -0.0847 -0.0708
( 78.83%) 0.8878*Br 7 s( 22.57%)p 3.43( 77.43%)
0.4751 -0.0033 -0.4319 -0.0109 0.7662
0.0230 0.0000 0.0000
4. (1.96807) BD ( 1)Al 1 -Br 8
( 21.17%) 0.4601*Al 1 s( 29.66%)p 2.28( 67.62%)d 0.09( 2.73%)
0.0000 -0.0001 0.5445 -0.0056 0.0001
0.4361 0.0324 0.0000 0.6941 0.0568
0.0000 0.0000 0.0000 0.1228 0.0000
0.0000 -0.0847 -0.0708
( 78.83%) 0.8878*Br 8 s( 22.57%)p 3.43( 77.43%)
0.4751 -0.0033 -0.4319 -0.0109 -0.7662
-0.0230 0.0000 0.0000
5. (1.96691) BD ( 1)Al 2 -Cl 3
( 12.14%) 0.3484*Al 2 s( 20.28%)p 3.77( 76.46%)d 0.16( 3.26%)
0.0000 0.0011 -0.4501 -0.0142 0.0003
-0.5362 -0.0191 0.0000 0.0000 0.0000
-0.0002 -0.6887 -0.0496 0.0000 -0.1524
0.0000 -0.0816 -0.0519
( 87.86%) 0.9374*Cl 3 s( 23.62%)p 3.23( 76.20%)d 0.01( 0.18%)
0.0000 0.0002 -0.4859 -0.0120 0.0000
0.7062 0.0012 0.0000 0.0000 0.0000
0.0000 0.5131 -0.0036 0.0000 -0.0356
0.0000 -0.0121 -0.0191
6. (1.96691) BD ( 1)Al 2 -Cl 4
( 12.14%) 0.3484*Al 2 s( 20.28%)p 3.77( 76.46%)d 0.16( 3.26%)
0.0000 0.0011 -0.4501 -0.0142 0.0003
-0.5362 -0.0191 0.0000 0.0000 0.0000
0.0002 0.6887 0.0496 0.0000 0.1524
0.0000 -0.0816 -0.0519
( 87.86%) 0.9374*Cl 4 s( 23.62%)p 3.23( 76.20%)d 0.01( 0.18%)
0.0000 0.0002 -0.4859 -0.0120 0.0000
0.7062 0.0012 0.0000 0.0000 0.0000
0.0000 -0.5131 0.0036 0.0000 0.0356
0.0000 -0.0121 -0.0191
7. (1.96807) BD ( 1)Al 2 -Br 5
( 21.17%) 0.4601*Al 2 s( 29.66%)p 2.28( 67.62%)d 0.09( 2.73%)
0.0000 -0.0001 0.5445 -0.0056 -0.0001
-0.4361 -0.0324 0.0000 0.6941 0.0568
0.0000 0.0000 0.0000 -0.1228 0.0000
0.0000 -0.0847 -0.0708
( 78.83%) 0.8878*Br 5 s( 22.57%)p 3.43( 77.43%)
0.4751 -0.0033 0.4319 0.0109 -0.7662
-0.0230 0.0000 0.0000
8. (1.96807) BD ( 1)Al 2 -Br 6
( 21.17%) 0.4601*Al 2 s( 29.66%)p 2.28( 67.62%)d 0.09( 2.73%)
0.0000 -0.0001 0.5445 -0.0056 -0.0001
-0.4361 -0.0324 0.0000 -0.6941 -0.0568
0.0000 0.0000 0.0000 0.1228 0.0000
0.0000 -0.0847 -0.0708
( 78.83%) 0.8878*Br 6 s( 22.57%)p 3.43( 77.43%)
0.4751 -0.0033 0.4319 0.0109 0.7662
0.0230 0.0000 0.0000
The bonding orbitals of Al, Cl and Br are sp3 hybridized.
References
1.[1]http://goldbook.iupac.org/CT07009.html 2.[2]http://science.jrank.org/pages/984/Bond-Energy.html
- ↑ 1.0 1.1 Cite error: Invalid
<ref>tag; no text was provided for refs namedhttp://goldbook.iupac.org/CT07009.html - ↑ 2.0 2.1 Cite error: Invalid
<ref>tag; no text was provided for refs namedhttp://science.jrank.org/pages/984/Bond-Energy.html