Rep:Mod:mb3210
Introduction
This computational chemistry course aims at giving an insight into bonding and chemical structure of molecules. Gaussview 5.0.9 linked to the Gaussian software was used to carry out calculations.
Week 1 consists of understanding the Gaussview 5.0.9 software and learning how to carry out calculations. This involves the optimisation, frequency analysis and molecular orbital analysis of a range of molecules, including BH3, TlBr3 and NH3. Kinetic and thermodynamic properties of the molecule can then be determined. When Gaussian carries out an optimisation, firstly, the nuclei are assumed to have fixed positions relative to the electrons (Born-Oppenheimer Approx), then the Schrödinger equation is solved with respect to the electrons, namely the SCF part of the calculation. Secondly, the Schrödinger equation is then solved for the position of the nuclei, namely the OPT part of the calculation (much more time consuming part of the calculation). The optimised molecule is the one with the lowest energy geometry from the calculations.
Week 2 is a mini-project where the techniques learnt from the previous week can be applied to further our understanding on the topic and cement our working knowledge of Gaussview 5.0. Additionally, the calculated data can be compared to experimental data as a way of judging the accuracy of experimental methods.
Optimising a molecule [Week 1]
Optimising a molecule of BH3
A borane molecule was created on Gaussview 5.0.9 with a trigonal planar structure and the bond lengths set to 1.5Å. The molecule was then optimised using a 3-21G basis set and the RB3LYP calculation method. The basis set determines the accuracy of the calculation, with 3-21G having very low accuracy but with the advantage of being a fast calculation. A 6-31G basis set for example is more accurate but more time consuming. The outcome is published below.
Optimised B-H bond distance: 1.19Å
Optimised H-B-H bond angle: 120.0°
| Parameters | |
|---|---|
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 3-21G |
| Charge | 0 |
| Spin | Singlet |
| Total energy(a.u.) | -26.46226338 |
| RMS Gradient(a.u.) | 0.00020672 |
| Dipole Moment(Debye) | 0.00 |
| Point Group | D3h |
| Time taken | 18.0 seconds |
Item Value Threshold Converged?
Maximum Force 0.000413 0.000450 YES
RMS Force 0.000271 0.000300 YES
Maximum Displacement 0.001610 0.001800 YES
RMS Displacement 0.001054 0.001200 YES
Predicted change in Energy=-1.071764D-06
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1935 -DE/DX = 0.0004 !
! R2 R(1,3) 1.1935 -DE/DX = 0.0004 !
! R3 R(1,4) 1.1935 -DE/DX = 0.0004 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
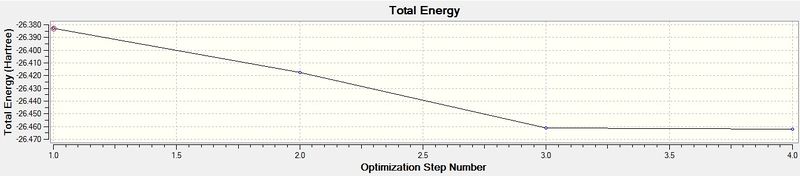
The above table is proof that the optimisation of the molecule was completed successfully, since it says "YES" under the "Converged?" heading. The RMS gradient is tending towards zero (less than 0.001), as seen in the graph below, which is also indicative of successful optimisation of a molecule, as well noticing that the B-H bond-length has decreased from the pre-set distance of 1.50Å, to an optimised distance of 1.19Å.
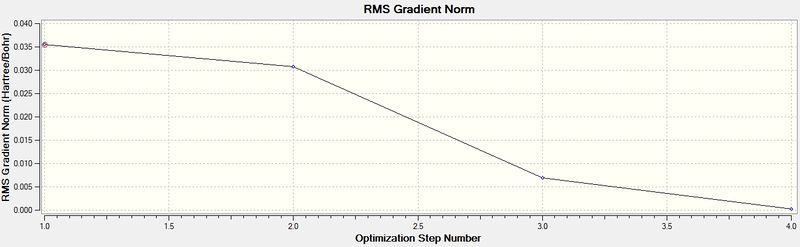
The energy graph is also shown below which further shows the optimisation tends to a more negative energy
The animation on the left show the optimisation structures. The structure where the B-H bond distance is at its smallest corresponds to the most negative energy in the optimisation plot and the smallest RMS gradient. Hence, this is the most stable structure found and is the optimised structure of the molecule. It will adopt this structure in the gas phase as well as in the solid state providing no solid state forces act to distort the molecule. It is also worth noticing that in the first step of the optimisation, the bonds connecting the atoms seem to disappear. However, this is not the case and the bonds between the atoms do still exist, rather the Gaussian software only draws bonds within a pre-defined distance separating the atoms. This is because in reality, bonds are not fixed lines joining two atoms, but instead should be viewed as an electrostatic force of attraction between the opposing charges of protons and electrons. Hence, a bond increases in strength as two atoms approach each other from infinite distance, not, as Gaussview suggests, that a bond is suddenly formed at a set distance. A potential energy curve of two atoms based on internuclear separation is a good illustration of this point.
Using a better basis set for BH3
The borane molecule optimised with the 3-21G basis set was further optimised using the more accurate 6-31G basis set. Again, the RB3LYP calculation method was applied. The results are published below.
Optimised B-H bond distance: 1.19Å
Optimised H-B-H bond angle: 120.0°
| Parameters | |
|---|---|
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Total energy(a.u.) | -26.61532363 |
| RMS Gradient(a.u.) | 0.00000235 |
| Dipole Moment(Debye) | 0.00 |
| Point Group | D3h |
| Time taken | 13.0 seconds |
Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.000019 0.001800 YES
RMS Displacement 0.000012 0.001200 YES
Predicted change in Energy=-1.304899D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1923 -DE/DX = 0.0 !
! R2 R(1,3) 1.1923 -DE/DX = 0.0 !
! R3 R(1,4) 1.1923 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
A difference in energy can be seen between the two basis sets used, however, we cannot compare them since they were computed using different basis sets. Only those calculations performed using the same basis sets and containing the same number of atoms can be compared.
Using pseudo-potentials and larger basis sets
Pseudo potentials: TlBr3
Heavy atoms such as thallium and bromine require the use of pseudo potentials which requires the basis set to be LanL2DZ. This is a medium level basis set where D95V is used on first row atoms and Los Alamos ECP on heavier elements. Once more, the RB3LYP calculation method was employed. One of the major advantages here of computing a molecule containing thallium, is we can understand some of its thermodynamic and kinetic properties without being exposed to the risks that thallium presents. The calculation method was set to RB3LYP and the point group constrained to D3H with a very tight tolerance (0.0001).
Optimised Tl-Br bond distance: 2.65Å
Optimised Br-Tl-Br bond angle: 120.0°
Dspace:http://hdl.handle.net/10042/22471
File:TlBr3 OPT LanL2DZ mb3210.LOG
| Parameters | |
|---|---|
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | LANL2DZ |
| Charge | 0 |
| Spin | Singlet |
| Total energy(a.u.) | -91.21812851 |
| RMS Gradient(a.u.) | 0.00000090 |
| Dipole Moment(Debye) | 0.00 |
| Point Group | D3h |
| Time taken | 34.9 seconds |
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000014 0.001200 YES
Predicted change in Energy=-6.084044D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.651 -DE/DX = 0.0 !
! R2 R(1,3) 2.651 -DE/DX = 0.0 !
! R3 R(1,4) 2.651 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
The optimisation animation of TlBr3 shows the structure where the bond lengths are optimised at the shortest distance to give the most negative energy and smallest RMS gradient.
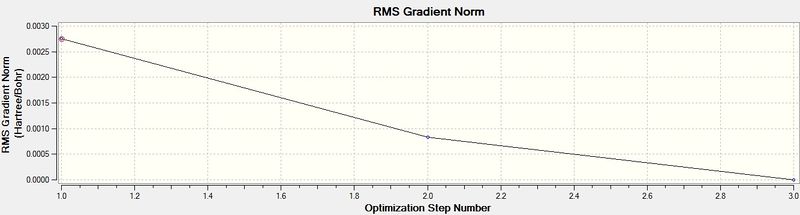
The energy graph is also shown below which further shows the optimisation tends to a more negative energy
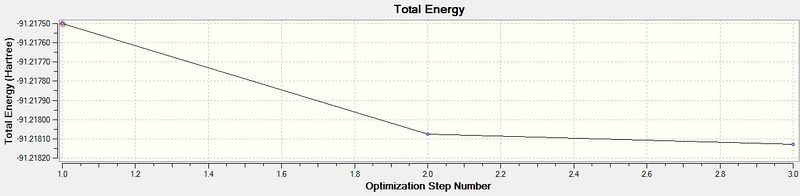
The calculated bond length is 2.65Å and experimentally the Tl-Br bond length in TlBr3 has been found to be 2.51Å.[1] This gives an error of 5.24% which can be attributed to the approximations used in the basis set for the calculations. A better basis set could have been chosen to optimise the bond lengths further, however, when selecting a basis set, it is important to consider whether choosing a better basis set outweighs a more time consuming process needed for the calculation.
Using a mixture of basis sets and pseudo-potentials: BBr3
The optimisation was carried out using the RB3LYP calculation method and the point group constrained to D3H with very tight tolerance (0.0001). The basis set used was Gen.

Optimised B-Br bond distance: 1.93Å
Optimised Br-B-Br bond angle: 120.0°
Dspace:http://hdl.handle.net/10042/22470
| Parameters | |
|---|---|
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | Gen |
| Charge | 0 |
| Spin | Singlet |
| Total energy(a.u.) | -64.43645296 |
| RMS Gradient(a.u.) | 0.00000382 |
| Dipole Moment(Debye) | 0.00 |
| Point Group | D3h |
| Time taken | 31.8 seconds |
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000036 0.001800 YES
RMS Displacement 0.000023 0.001200 YES
Predicted change in Energy=-4.026911D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.934 -DE/DX = 0.0 !
! R2 R(1,3) 1.934 -DE/DX = 0.0 !
! R3 R(1,4) 1.934 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------

As before, the optimisation animation shows the shortest bond length which gives an optimised structure to give the most negative energy and the smallest RMS gradient.

The energy graph is also shown below which further shows the optimisation tends to a more negative energy

Comparison of computed bond lengths on optimised structures of BH3 , TlBr3 and BBr3
| Bond | Calculated Bond Length (Å) | Experimental Bond Length (Å) | Basis Set |
|---|---|---|---|
| B-H | 1.19 | 1.08 (in hexamethylenetetramine-borine)[2] | 6-31G(d,p) |
| Tl-Br | 2.65 | 2.51[1] | LanL2DZ |
| B-Br | 1.93 | 2.01[3] | Gen |
The above data shows how bond lengths differ depending on the size of the atoms involved, and hence how the orbital overlap varies. Comparison of the B-H bond (1.19Å) with the B-Br bond (1.93Å) shows that the presence of the larger Br atom compared to H results in a much longer bond. Despite the greater electronegativity difference in B-Br than B-H, there is poorer orbital overlap in B-Br since the size of the atoms differ greatly and the Br orbitals are more diffuse. In B-H however, the atoms are of similar size, so the overlap is greater. Hence, the bond in B-Br is weaker and therefore longer.
Likewise, a comparison can be made between the Tl-Br bond length (2.65Å) and that of B-Br (1.93Å). Thallium is in the same group as Boron, but four periods below. Therefore, a useful comparison can be drawn by examining the effect on bond length when changing the Group 13 atom. Br is two periods above Tl and two periods below B. Tl is a large atom in the 6th period of the periodic table, hence its valence shell is far more diffuse than that of Boron. Therefore, the overlap of the orbitals in B and Br is far greater than that in Tl and Br. As a result, the B-Br bond is shorter and stronger than Tl-Br.
Experimental data shows similar bond lengths to those calculated and limitations of experimental techniques can account for the differences.
Frequency analysis
BH3
| Parameters | |
|---|---|
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Total energy(a.u.) | -26.61532363 |
| RMS Gradient(a.u.) | 0.00000237 |
| Dipole Moment(Debye) | 0.00 |
| Point Group | D3h |
| Time taken | 11.0 seconds |
Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000002 0.000300 YES
Maximum Displacement 0.000019 0.001800 YES
RMS Displacement 0.000009 0.001200 YES
Predicted change in Energy=-1.323374D-10
Optimization completed.
-- Stationary point found.
Low frequencies --- -0.9033 -0.7343 -0.0054 6.7375 12.2491 12.2824 Low frequencies --- 1163.0003 1213.1853 1213.1880
We know there should be 3N-6 vibrational frequencies present. For BH3, N=4 so we can expect 6 vibrations as the frequency table shows below. However, in the infrared spectrum also shown below, only 3 peaks are present. This can be explained by considering the symmetry of the vibrations. Firstly. vibrational mode 4 is totally symmetric, displaying A'1 symmetry. As a result it is infrared inactive. From the remaining 5 vibrational modes, there is one pair of degenerate vibrations at 1213 cm-1 with E' symmetry, one pair at 2715 cm-1 also with E' symmetry and one mode at 1163 cm-1 with A' '2 symmetry. Hence, there are three distinct vibrations.

| No. | Form of vibration | Frequency(cm-1) | Intensity | Symmetry D3h point group |
|---|---|---|---|---|
| 1 | All H atoms move together in a concerted motion up and down in the plane of the principal axis. The B atom moves up and down the same plane, but in the opposite direction to the H atoms. This causes a distinct change in dipole moment | 1163 | 93 | A' '2 |
| 2 | One of the H atoms remains in a fixed position in relation to the B atom, while the other two H atoms bend towards each other in the plane perpendicular to the principal axis (In-plane scissoring) | 1213 | 14 | E' |
| 3 | For two hydrogens, the H-B-H bond angle remains constant and the atoms bend clockwise together. The third H atom moves towards and away from each other H atom in turn (Asymmetric scissoring) | 1213 | 14 | E' |
| 4 | B atom stationary, all H atoms move in and out in the plane of a B-H bond together in a concerted motion (In-plane symmetric stretching) | 2582 | 0 | A '1 |
| 5 | For one H atom, the B-H bond remains fixed, while for the other two H atoms the B-H bond shortens and lengthens equal and opposite amounts to each other (Asymmetric stretching) | 2715 | 126 | E' |
| 6 | For two H atoms, the B-H bonds shorten and lengthen equally together, while the other B-H bond lengthens when the other two B-H bonds shorten and vice versa (2 hydrogens stretching symmetrically in-phase, one stretching of out phase) | 2715 | 126 | E' |
| Frequency vibration no. | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |||||||||||||||||||
| Animation |
|
|
|
|
|
| ||||||||||||||||||
TlBr3
Dspace:http://hdl.handle.net/10042/22450
| Parameters | |
|---|---|
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | LANL2DZ |
| Charge | 0 |
| Spin | Singlet |
| Total energy(a.u.) | -91.21812851 |
| RMS Gradient(a.u.) | 0.00000088 |
| Imaginary Freq | 0 |
| Dipole Moment(Debye) | 0.00 |
| Point Group | D3h |
| Time taken | 25.2 seconds |
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000011 0.001200 YES
Predicted change in Energy=-5.660901D-11
Optimization completed.
-- Stationary point found.
Low frequencies --- -3.4213 -0.0026 -0.0004 0.0015 3.9367 3.9367 Low frequencies --- 46.4289 46.4292 52.1449
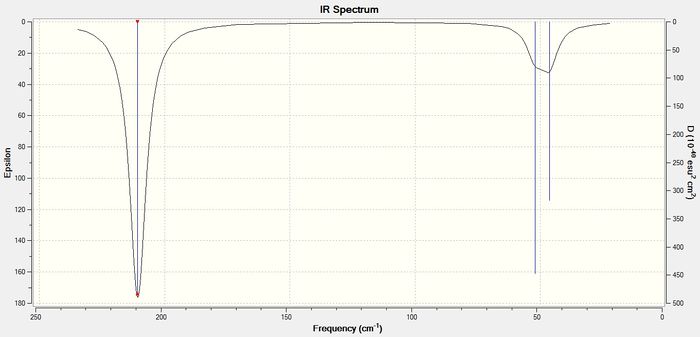
Similarly to BH3, the infrared spectrum of TlBr3 shows 3 peaks. These are at 46 cm-1 (lowest "real" normal mode), which corresponds to vibrational modes 1 and 2, 52 cm-1 corresponding to vibration 3 and at 211 cm-1 which represents vibrations 5 and 6. Vibrational mode 4 has no peak in the infrared spectrum since it is totally symmetric and therefore infrared inactive.
| No. | Form of vibration | Frequency(cm-1) | Intensity | Symmetry D3h point group |
|---|---|---|---|---|
| 1 | One of the Br atoms remains in a fixed position in relation to the Tl atom, while the other two Br atoms bend towards each other in the plane perpendicular to the principal axis (In-plane scissoring) | 46 | 4 | E' |
| 2 | For two hydrogens, the Br-Tl-Br bond angle remains constant and the atoms bend clockwise together. The third Br atom moves towards and away from each other Br atom in turn (Asymmetric scissoring) | 46 | 4 | E' |
| 3 | All Br atoms move together in a concerted motion up and down in the plane of the principal axis. The Tl atom moves up and down the same plane, but in the opposite direction to the Br atoms. This causes a distinct change in dipole moment | 52 | 6 | A' '2 |
| 4 | Tl atom stationary, all Br atoms move in and out in the plane of a Tl-Br bond together in a concerted motion (In-plane symmetric scissoring) | 165 | 0 | A '1 |
| 5 | For one Br atom, the Tl-Br bond remains fixed, while for the other two H atoms the Tl-Br bond shortens and lengthens equal and opposite amounts to each other (Asymmetric scissoring) | 211 | 25 | E' |
| 6 | For two Br atoms, the Tl-Br bonds shorten and lengthen equally together, while the other Tl-Br bond lengthens when the other two Tl-Br bonds shorten and vice versa (2 bromine atoms stretching symmetrically in-phase, one stretching of out phase) | 211 | 25 | E' |
| Frequency vibration no. | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |||||||||||||||||||
| Animation |
|
|
|
|
|
| ||||||||||||||||||
Comparison of vibrational frequencies of TlBr3 and TlBr3
| Vibrational frequencies (cm-1) | |
|---|---|
| TlBr3 | BH3 |
| 46 | 1163 |
| 46 | 1213 |
| 52 | 1213 |
| 165 | 2582 |
| 211 | 2715 |
| 211 | 2715 |
The first obvious comparison to be made is that the vibrational frequencies in BH3 are two orders of magnitude larger than in TlBr3. This can be attributed to the greater bond strength in the B-H bond in comparison to the Tl-Br bond and is in accordance with the significantly shorter B-H bond length. We can invoke the same argument as used in the comparison of bond lengths. The more diffuse orbitals in the Tl-Br bond results in poorer orbital overlap, a weaker bond and hence a lower vibrational frequency. Another approach to explaining the lower vibrational frequency in TlBr3 is to consider Hooke's Law. This relates the vibrational frequency of a molecule to its redcued mass. Hence, the larger the two atoms, the greater their reduced mass, so the lower their vibrational frequency. It is worth noting that whilst each molecule has 6 vibrational modes, the ordering differs slightly between the two. Similarly, the infrared spectra for each molecule show three peaks, relating to the two being isostructural. In BH3, the ordering is A' ' 2, E', E', A'1, E', E' whereas in TlBr3 it is E', E', A' '2, A'1, E', E'. However, for both, the A'1 and E' vibrations are close in energy, while the A' '2 and the other set of E' vibrations are also energetically close. In BH3, the first 3 modes are bending frequencies whilst the other 3 modes are stretching frequencies which could explain the similarities in their vibrational frequencies. The same can be said for TlBr3.
It is important to note that if any comparison is to be attempted between calculations that they use the same method basis set for optimisation and frequency analysis. Calculations using a different basis set are not comparable and no conclusions can be drawn. The purpose of carrying out a frequency analysis is to confirm that the optimised structure obtained is a minimum. It is also very useful in analysing and understanding the vibrational modes in molecules and helps in the interpretation of infrared spectra. The "low frequencies" found in the .log file represent the "-6" part in the equation for the number of vibrational modes, "3N-6", and are representative of the motions of the centre of mass of the molecule.
Molecular orbital analysis
Population analysis of BH3
Dspace:http://hdl.handle.net/10042/22469
| Parameters | |
|---|---|
| File Type | .log |
| Calculation Type | SP |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Total energy(a.u.) | -26.61532363 |
| Dipole Moment(Debye) | 0.00 |
| Point Group | D3h |
| Time taken | 16 minutes 21.0 seconds |
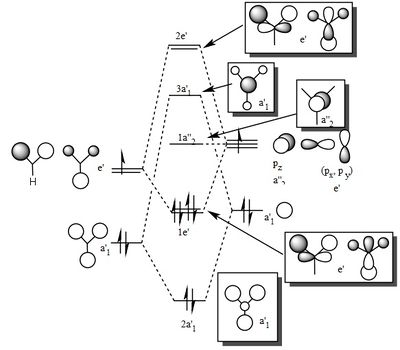
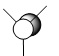
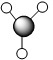
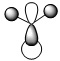
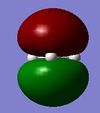
| Method | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| LCAO |  |
 |
 |
 |
 |
 |

|
| Real |  |
 |
 |
 |
 |
 |

|
| Symmetry label | a'1 | e' | e' | a2 | a'1 | e' | e' |
From the comparison table above, it is easy to see that the LCAO method for constructing MOs produces a very good fit in comparison to the calculated "Real" MOs in Gaussview. Hence, the LCAO approach proves that qualitative MO theory can be used for accurate modelling of MOs for small, simple molecules.
NBO Analysis
NH3
Optimisation
The optimisation of NH3 was carried out in Gaussview using the RB3LYP calculation method and a 6-31G basis set, applying the "nosymm" keyword. The summary data is shown below.
| Parameters | |
|---|---|
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Total energy(a.u.) | -56.55776856 |
| RMS Gradient(a.u.) | 0.00000885 |
| Dipole Moment(Debye) | 1.85 |
| Point Group | C1 |
| Time taken | 18.0 seconds |
Optimised N-H bond distance: 1.02Å
Optimised H-N-H bond angle: 105.7°
Item Value Threshold Converged?
Maximum Force 0.000024 0.000450 YES
RMS Force 0.000012 0.000300 YES
Maximum Displacement 0.000079 0.001800 YES
RMS Displacement 0.000053 0.001200 YES
Predicted change in Energy=-1.629728D-09
Optimization completed.
-- Stationary point found.
Frequency analysis
| Parameters | |
|---|---|
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Total energy(a.u.) | -56.55776856 |
| RMS Gradient(a.u.) | 0.00000878 |
| Dipole Moment(Debye) | 1.85 |
| Point Group | C1 |
| Time taken | 9.0 seconds |
Item Value Threshold Converged?
Maximum Force 0.000007 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.000021 0.001800 YES
RMS Displacement 0.000009 0.001200 YES
Predicted change in Energy=-6.353265D-11
Optimization completed.
-- Stationary point found.
Low frequencies --- -31.6906 -0.0009 0.0003 0.0006 13.0070 25.5725 Low frequencies --- 1089.5009 1694.1085 1694.1682
Population analysis
| Parameters | |
|---|---|
| File Type | .log |
| Calculation Type | SP |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Total energy(a.u.) | -56.55776856 |
| Dipole Moment(Debye) | 1.85 |
| Point Group | C1 |
| Time taken | 8 minutes 8.3 seconds |
NBO analysis of NH3
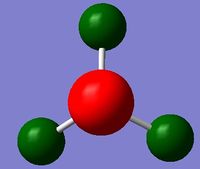

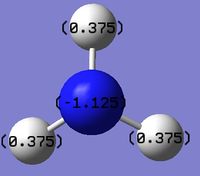
The charge range can be seen above and is -1.125 to +1.125
The NBO charges were then shown using Gaussview:
Nitrogen NBO charge = -1.125
Hydrogen NBO charge = +0.375
NH3BH3 dissociation energy analysis
Optimisation
3-21G basis set
The initial optimisation was carried out using a 3-21G basis set and RB3LYP calculation method.
Dspace:http://hdl.handle.net/10042/22502
File:NH3BH3 OPT 321G MB3210.log
| Parameters | |
|---|---|
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 3-21G |
| Charge | 0 |
| Spin | Singlet |
| Total energy(a.u.) | -82.76661837 |
| RMS Gradient(a.u.) | 0.00003006 |
| Dipole Moment(Debye) | 5.84 |
| Point Group | C1 |
| Time taken | 59.4 seconds |
Item Value Threshold Converged?
Maximum Force 0.000094 0.000450 YES
RMS Force 0.000030 0.000300 YES
Maximum Displacement 0.000419 0.001800 YES
RMS Displacement 0.000178 0.001200 YES
Predicted change in Energy=-5.742846D-08
Optimization completed.
-- Stationary point found.
6-31G(d,p) basis set
The optimisation was then carried out using the more accurate 6-31G(d,p) basis set and a RB3LYP calculation method.
Dspace:http://hdl.handle.net/10042/22503
File:NH3BH3 OPT 631G MB3210.log
| Parameters | |
|---|---|
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Total energy(a.u.) | -83.22469001 |
| RMS Gradient(a.u.) | 0.00005640 |
| Dipole Moment(Debye) | 5.56 |
| Point Group | C1 |
| Time taken | 59.1 seconds |
Item Value Threshold Converged?
Maximum Force 0.000132 0.000450 YES
RMS Force 0.000037 0.000300 YES
Maximum Displacement 0.001235 0.001800 YES
RMS Displacement 0.000549 0.001200 YES
Predicted change in Energy=-1.183236D-07
Optimization completed.
-- Stationary point found.
Frequency analysis
Dspace:http://hdl.handle.net/10042/22504
| Parameters | |
|---|---|
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Total energy(a.u.) | -83.22469001 |
| RMS Gradient(a.u.) | 0.00005643 |
| Dipole Moment(Debye) | 5.56 |
| Point Group | C1 |
| Time taken | 57.0 seconds |
Item Value Threshold Converged?
Maximum Force 0.000254 0.000450 YES
RMS Force 0.000056 0.000300 YES
Maximum Displacement 0.001414 0.001800 YES
RMS Displacement 0.000702 0.001200 YES
Predicted change in Energy=-2.149394D-07
Optimization completed.
-- Stationary point found.
Low frequencies --- -0.0011 -0.0008 0.0008 6.6382 14.5267 20.1742 Low frequencies --- 263.3505 631.3096 638.0440
Dissociation energy
E(NH3): -56.55776856
E(BH3): -26.61532363
E(NH3BH3): -83.22469001
Energy difference: ΔE=E(NH3BH3)-[E(NH3)+E(BH3)]
Hence, ΔE= (-83.22469001)-[(-56.55776856)+(-26.61532363)]
ΔE= -0.05159782 a.u.
Therefore, Edissociation= -135.47 kJmol-1
Mini Project [Week 2] - Aromaticity
All calculations hereafter were performed on the HPC servers using a calculation method of RB3LYP and a 6-31G(d,p) basis set.
Benzene
Optimisation

Dspace: http://hdl.handle.net/10042/22512
File:Benzene OPT 631G mb3210.log
| Parameters | |
|---|---|
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Total energy(a.u.) | -232.25821387 |
| RMS Gradient(a.u.) | 0.00008450 |
| Dipole Moment(Debye) | 0.00 |
| Point Group | D6H |
| Time taken | 1 minute 3.7 seconds |
Optimised C-C bond length: 1.40Å
Optimised C-H bond length: 1.09Å
Item Value Threshold Converged?
Maximum Force 0.000199 0.000450 YES
RMS Force 0.000081 0.000300 YES
Maximum Displacement 0.000847 0.001800 YES
RMS Displacement 0.000299 0.001200 YES
Predicted change in Energy=-4.636830D-07
Optimization completed.
-- Stationary point found.
Frequency analysis
Dspace: http://hdl.handle.net/10042/22513
File:Benzene FREQ mb3210.log
| Parameters | |
|---|---|
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Total energy(a.u.) | -232.25821387 |
| RNS Gradient(a.u.) | 0.00008452 |
| Imaginary Freq | 1 |
| Dipole Moment(Debye) | 0.00 |
| Point Group | D6H |
| Time taken | 1 minute 28.9 seconds |
Item Value Threshold Converged?
Maximum Force 0.000199 0.000450 YES
RMS Force 0.000085 0.000300 YES
Maximum Displacement 0.000806 0.001800 YES
RMS Displacement 0.000346 0.001200 YES
Predicted change in Energy=-4.349367D-07
Optimization completed.
-- Stationary point found.
Low frequencies --- -4.6046 -4.6046 -0.0088 -0.0043 -0.0042 9.6590 Low frequencies --- 413.9386 413.9386 621.1428
Population analysis
Dspace: http://hdl.handle.net/10042/22540
File:Benzene NBO mb3210.log
| Parameters | |
|---|---|
| File Type | .log |
| Calculation Type | SP |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Total energy(a.u.) | -232.25821387 |
| Dipole Moment(Debye) | 0.00 |
| Point Group | D6H |
| Time taken | 54.1 seconds |
MO diagram
The benzene molecular orbital diagram shown below shows the "centre" part of the molecular orbitals present in benzene, ranging from MOs 7 to 32 (7-21 occupied, 22-32 unoccupied) in Gaussview. The MO diagram starts with all bonding s orbitals, going through to all bonding p orbitals, finishing with anti-bonding generate MOs composed of s orbitals. There are many more MOs calculated in Gaussview, however, these soon become very complex to interpret and move away from the LCAO. The symmetry of each MO was determined to assist in identifying degernate MOs (can be found in the .log file).
The concept of aromaticity can be easily represented in benzene. For a molecule to be aromatic, it must follow Hückel's rule that an aromatic molecule contains 4n+2 π electrons, where n is an integer. Benzene has 6π electrons, hence n=2, so benzene conforms to Hückel's rule. Likewise, both pyridinium and boratabenzene also contain 6π electrons due to their respective charges offsetting the difference in valence electrons between carbon and both nitrogen and boron. Hence, they too are aromatic molecules. Pyridinium derivatives, such as pyridinium chlorochromate, have beneficial uses, being used as a mild oxidating agent for converting alcohols to carbonyl compounds.[4] Borazine, on the other hand, has a long history focusing on its aromaticity, being the subject of much debate. It can be described as a π aromatic compound, but it is not globally aromatic as the electronic system is not as delocalised as in benzene.[5] Therefore, it is still considered aromatic and is often referred to as "inorganic benzene".[5]
Pyridinium
Optimisation
Dspace: http://hdl.handle.net/10042/22515
File:Pyridinium OPT 631G mb3210.log
| Parameters | |
|---|---|
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 1 |
| Spin | Singlet |
| Total energy(a.u.) | -248.66807395 |
| RMS Gradient(a.u.) | 0.00003889 |
| Dipole Moment(Debye) | 1.87 |
| Point Group | C2V |
| Time taken | 1 minute 52.4 seconds |
Optimised N-C bond length: 1.35Å
Optimised N-H bond length: 1.02Å
Item Value Threshold Converged?
Maximum Force 0.000063 0.000450 YES
RMS Force 0.000023 0.000300 YES
Maximum Displacement 0.000698 0.001800 YES
RMS Displacement 0.000178 0.001200 YES
Predicted change in Energy=-7.097421D-08
Optimization completed.
-- Stationary point found.
Frequency analysis
Dspace: http://hdl.handle.net/10042/22516
File:Pyridinium FREQ mb3210.log
| Parameters | |
|---|---|
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 1 |
| Spin | Singlet |
| Total energy(a.u.) | -248.66807395 |
| RMS Gradient(a.u.) | 0.00003891 |
| Dipole Moment(Debye) | 1.87 |
| Point Group | C2V |
| Time taken | 2 minutes 27.7 seconds |
Item Value Threshold Converged?
Maximum Force 0.000155 0.000450 YES
RMS Force 0.000039 0.000300 YES
Maximum Displacement 0.000758 0.001800 YES
RMS Displacement 0.000231 0.001200 YES
Predicted change in Energy=-7.607867D-08
Optimization completed.
-- Stationary point found.
Low frequencies --- -7.2529 -0.0006 0.0009 0.0010 17.3499 18.5758 Low frequencies --- 392.4576 404.0624 620.4718
Population analysis
Dspace: http://hdl.handle.net/10042/22541
File:Pyridinium NBO mb3210.log
| Parameters | |
|---|---|
| File Type | .log |
| Calculation Type | SP |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 1 |
| Spin | Singlet |
| Total energy(a.u.) | -248.66807395 |
| Dipole Moment(Debye) | 1.87 |
| Point Group | C2V |
| Time taken | 58.1 seconds |
Boratabenzene
Optimisation
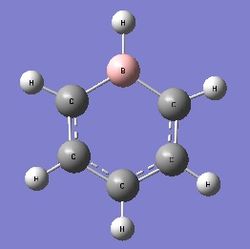
Dspace: http://hdl.handle.net/10042/22517
File:Boratabenzene OPT 631G mb3210.log
| Parameters | |
|---|---|
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | -1 |
| Spin | Singlet |
| Total energy(a.u.) | -219.02052980 |
| RMS Gradient(a.u.) | 0.00016432 |
| Dipole Moment(Debye) | 2.85 |
| Point Group | C2V |
| Time taken | 1 minute 54.6 seconds |
Optimised B-H bond length: 1.22Å
Optimised B-C bond length: 1.51Å
Item Value Threshold Converged?
Maximum Force 0.000158 0.000450 YES
RMS Force 0.000071 0.000300 YES
Maximum Displacement 0.000914 0.001800 YES
RMS Displacement 0.000329 0.001200 YES
Predicted change in Energy=-6.959555D-07
Optimization completed.
-- Stationary point found.
Frequency analysis
Dspace: http://hdl.handle.net/10042/22518
File:Boratabenzene FREQ mb3210.log
| Parameters | |
|---|---|
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | -1 |
| Spin | Singlet |
| Total energy(a.u.) | -219.02052979 |
| RMS Gradient(a.u.) | 0.00016443 |
| Dipole Moment(Debye) | 2.85 |
| Point Group | C2V |
| Time taken | 2 minutes 30.3 seconds |
Item Value Threshold Converged?
Maximum Force 0.000434 0.000450 YES
RMS Force 0.000164 0.000300 YES
Maximum Displacement 0.000889 0.001800 YES
RMS Displacement 0.000398 0.001200 YES
Predicted change in Energy=-7.654683D-07
Optimization completed.
-- Stationary point found.
Low frequencies --- -13.1151 0.0004 0.0009 0.0009 15.0343 18.1624 Low frequencies --- 371.3435 404.2381 565.2603
Population analysis
Dspace: http://hdl.handle.net/10042/22542
File:Boratabenzene NBO mb3210.log
| Parameters | |
|---|---|
| File Type | .log |
| Calculation Type | SP |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | -1 |
| Spin | Singlet |
| Total energy(a.u.) | -219.02052979 |
| Dipole Moment(Debye) | 2.85 |
| Point Group | C2V |
| Time taken | 57.9 seconds |
Borazine
Optimisation
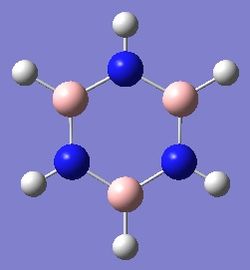
Dspace: http://hdl.handle.net/10042/22548
File:Borazine OPT 631G mb3210.log
| Parameters | |
|---|---|
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Total energy(a.u.) | -242.68458184 |
| RMS Gradient(a.u.) | 0.00006419 |
| Dipole Moment(Debye) | 0.00 |
| Point Group | D3H |
| Time taken | 2 minutes 28.6 seconds |
Item Value Threshold Converged?
Maximum Force 0.000085 0.000450 YES
RMS Force 0.000033 0.000300 YES
Maximum Displacement 0.000251 0.001800 YES
RMS Displacement 0.000075 0.001200 YES
Predicted change in Energy=-9.320688D-08
Optimization completed.
-- Stationary point found.
Optimised B-N bond length: 1.43Å
Optimised B-H bond length: 1.19Å
Optimised N-H bond length: 1.01Å
Experimentally, the B-N bond length is 1.44Å[6] compared to the calculated length of 1.43Å (0.7% error), the B-H bond length was found to be 1.20Å[6] compared to the calculated length of 1.19Å(0.83% error) and the N-H bond length was found to be 1.02Å[6] compared to the optimised length of 1.01Å(0.99% error). These bond lengths show that literature values obtained experimentally and calculated values are in very close agreement, emphasising the accuracy of experimental techniques as well as showing that Gaussview combined with Gaussian are very powerful tools in modern chemistry.
Frequency analysis
Dspace: http://hdl.handle.net/10042/22550
File:Borazine FREQ mb3210.log
| Parameters | |
|---|---|
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Total energy(a.u.) | -242.68458184 |
| RMS Gradient(a.u.) | 0.00006436 |
| Imaginary Freq | 0 |
| Dipole Moment(Debye) | 0.00 |
| Point Group | D3H |
| Time taken | 2 minutes 1.3 seconds |
Item Value Threshold Converged?
Maximum Force 0.000203 0.000450 YES
RMS Force 0.000064 0.000300 YES
Maximum Displacement 0.000311 0.001800 YES
RMS Displacement 0.000098 0.001200 YES
Predicted change in Energy=-1.075851D-07
Optimization completed.
-- Stationary point found.
Low frequencies --- -12.7010 -12.4894 -9.0645 -0.0099 0.0374 0.0776 Low frequencies --- 289.1171 289.1263 403.9186
Population analysis
Dspace: http://hdl.handle.net/10042/22551
File:Borazine NBO mb3210.log
| Parameters | |
|---|---|
| File Type | .log |
| Calculation Type | SP |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Total energy(a.u.) | -242.68458184 |
| Dipole Moment(Debye) | 0.00 |
| Point Group | D3H |
| Time taken | 51.9 seconds |
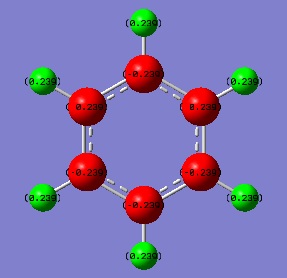
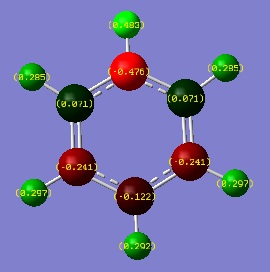
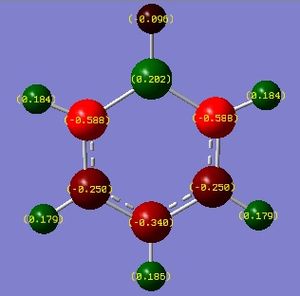
Comparing NBO analysis of benzene, pyridinium, boratabenzene and borazine
Links to Dspace for the NBO analysis can be found under "Population analysis" section of each molecule.
Comparison of molecular orbitals
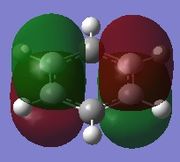
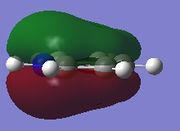
| Molecule | Fully s-bonding | π1 | HOMO |
|---|---|---|---|
| Benzene | 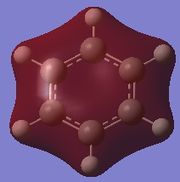 |
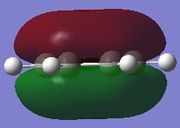 |
|
| Pyridinium |  |
 |
 |
| Boratabenzene | 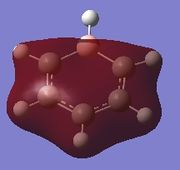 |
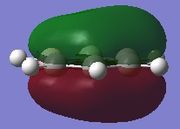 |
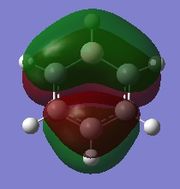 |
| Borazine |  |
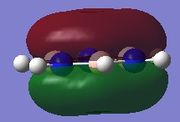 |
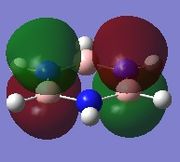 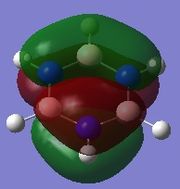
|
Fully s-bonding
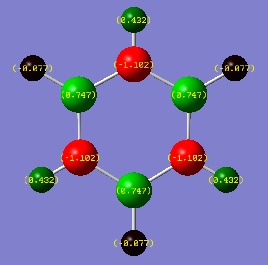
Since this is the fully bonding s orbital molecular orbitsl there are no nodal planes present. Benzene will be used as a reference molecule. It is totally symmetric and has an even, delocalised electron distribution amongst its carbon atoms. Borazine, similar to benzene is highly symmetric, however there is a slight difference in the electron distribution due to the difference in electronegativity of nitrogen and boron (Pauling electronegativity values, N = 3.0, B = 2.0)[7][8]. It can be seen that there is reduced electron density at the boron atoms and increased electron density at the nitrogen atoms. The presence of the nitrogen atom in pyridinium distorts the electron cloud towards the nitrogen where there is greater electron density compared to the carbon atoms in the molecule. In contrast, in boratabenzene, since it is less electronegative than carbon, there is more electron density localised on the carbon atoms in the ring making the boron atom electron deficient.
π1
The π1 molecular orbital for each molecule demonstrates the concept of electron delocalisation, since there is a region of electron density delocalised above and below the plane of the molecule, with a nodal plane perpendicular to the principal axis. Hence, this illustrates that all four molecules are aromatic. On closer inspection in can be seen however, that the distribution of the electron density in both pyridinium and boratabenzene is not uniform as seen in both benzene and borazine. This is due to the difference in electronegativity between the heteroatom and the carbon atoms. In boratabenzene, the electron density appears to be deficient on the boron atom since it is electropostive with respect to carbon (Pauling electronegativity values, B = 2.0, C = 2.5[8]). Therefore, the calculated MO concurs with what we would expect to observe and the boron atom can be considered to be donating less electron density into the π1 molecular orbital compared to carbon. As a result, using benzene as a reference, boratabenzene can be considered to be less stable that benzene due to the Lewis acidity of the boron atom, which would disrupt the aromaticity of the molecule. On the other hand, in pyridinium, the difference in Pauling electronegativities in nitrogen and carbon results in more electron density being localised on the more electronegative nitrogen atom compared to the carbon atoms (Pauling electronegativity values, N = 3.0, C = 2.5[7]. Therefore, the nitrogen atom donates more electron density into the π1 molecular orbital compared to carbon. As a result pyridinium is more stable than benzene since the lone pair on nitrogen is perpendicular to the aromatic ring so doe not contributre to the aromatic system. This means that protonation of nitrogen does not disrupt the aromaticity giving a more stable molecule than benzene, which when protonated loses its aromaticity.
HOMO
By analysing the energy of the individual MOs produced in GaussView, it is apparent that both benzene and borazine have degenerate HOMOs corresponding to π2 + π3, while both pyridinium and boratabenzene have only one MO in the HOMO, π2. This can be attributed to the higher degree of symmetry found in both benzene and borazine compared to pyridinium and boratabenzene, where a carbon atom has been substituted for a heteroatom. The HOMOs across all four molecules have a nodal plane in the plane of the principal axis. For benzene and borazine, one of the MOs has 4 pz orbitals, 2 in phase on one side of the ring, 2 out of phase on the other side, while the other MO has 6 pz orbitals with 3 in phase on one side of the ring and 3 out of phase on the other side of the ring. In pyridinium, the HOMO has 4 pz orbitals, 2 in phase on one side of the ring, 2 out of phase on the other side. In boratabenzene, the HOMO contains 6 pz orbitals with 3 in phase on one side of the ring and 3 out of phase on the other side of the ring.
Comparison of MO energies between benzene, pyridinium, boratabenzene and borazine
| Molecule | Fully s-bonding | π1 | HOMO |
|---|---|---|---|
| Benzene | -0.84678 | -0.35999 | -0.24691 (degenerate) |
| Pyridinium | -1.21402 | -0.64064 | -0.47885 |
| Boratabenzene | -0.60434 | -0.13208 | +0.01093 |
| Borazine | -0.88851 | -0.36130 | -0.27590 (degenerate) |
Fully s-bonding
The fully bonding s-orbitals for each molecule represents the lowest energy all bonding combination of orbitals. There are orbitals much lower in energy in each molecule, however, these consist of the 1s orbitals for a couple of the carbon atoms, and therefore do not represent a bonding combination. The energies generated help emphasise the point noted earlier which was that pyridinium is more stable than benzene, while boratabenzene is less so. The energy for pyridinium is more negative than benzene, indicating the greater stability of this MO, while for boratabenzene it is less negative. Borazine has a similar energy to benzene, highlighting their similarities in structure.
π1
Again, the same trends can be seen as noted for the fully s-bonding case. The energies for π1 are less negative than the fully s-bonding case since the overlap of p orbitals is not as great as in s orbitals, resulting in a slightly less stable molecular orbital.
HOMO
Once more, the same trends are encountered and likewise the same argument can be invoked. It is worth highlighting, however, that the HOMO for boratabenzene is positive in energy, indicating its relative instability compared to benzene's HOMOs which are negative. This further emphasises the high Lewis acidity of the boron atom in boratabenzene, which favours the formation of adducts to stablise the molecule.
Effects on the MO diagrams
Benzene and borazine will exhibit similar molecular orbital diagrams due to their high symmetry which can be seen via the presence of degenerate MOs found in found molecules. Slight differences may arise due to the differing orbital sizes and hence overlap of the orbitals, resulting in differing stabilities. For boratabenzene and pyridinium, however, we can expect more noticeable changes which will arise, due to the electronegativity difference of the heteroatoms with carbon and the loss of degeneracy associated with this. This will effect the ordering of the MOs and as a result it can be expected that boratabenzene and pyridinium will not show the same ordering.
Conclusion
This inorganic computation chemistry course has highlighted the importance and usefulness of modern computational chemistry. It has been demonstrated to be a powerful technique of which experimental data may be compared to, since a vast number of molecules known today can be optimised, frequency analysis undertaken and the computation of its molecular orbitals can be completed. As a result, our understanding of chemistry at the molecular level has improved, gaining a greater working knowledge of the reactivity of molecules and has enabled further advances in this field of chemistry. In terms of this project we have optimised a number of molecules and carried out a frequency analysis on them as well as a population analysis. We have shown the similarities of benzene and borazine in terms of their molecular orbitals and energy associated with them, as well as showing how the substitution of a carbon atom in benzene for a heteroatom can disrupt the degeneracy of the molecule.
References
- ↑ 1.0 1.1 J. Glaser, G. Johansson, "On the Structures of the Hydrated Thallium(III) Ion and its Bromide Complexes in Aqueous Solution", Acta Chem. Scan., 1992, 36, 125.
- ↑ F. Hanic, V. Šubrtová, "The Crystal Structure of Hexamethylenetetramine-Borine, C6H12N4.BH3", Acta Cryst., 1969, B25, 405.
- ↑ F. Allen, L. Brammer, R. Taylor, "Tables of Bond Lengths determined by X-Ray and Neutron Diffraction. Part 1. Bond Lengths in Organic Compounds", J. Chem. Soc., 1987, S5.
- ↑ E.F.V. Scriven, J. Toomey, R. Murugan, "Pyridine and Pyridine Derivatives", Kirk-Othmer Ency. Chem. Tech., 2000, 7
- ↑ 5.0 5.1 R. Islas, E. Chamorro, J. Robles, T. Heine, J.C. Santos, G. Merino, "Borazine: to be or not to be aromatic", Struct. Chem., 2007, 1-8
- ↑ 6.0 6.1 6.2 V.V. Volkov, K.G. Myakishev, E.A. Ilinchik, "Borazine and its derivatives: Synthesis, Mechanochemical Synthesis Reactions and Outlooks for Technological Applications", Chem. for Sustainable Development, 2009, 227
- ↑ 7.0 7.1 J.O McCaldin, T.C. McGill, C. A. Mead, "Correlation for III-V and II-VI Semiconductors of the Au Schottky Barrier Energy with Anion ELectronegativity", Phys. Rev. Lett., 1976, 36, 56-58
- ↑ 8.0 8.1 G. L. Krasko, "Effect of Boron, Carbon, Phosphorus and Sulfur on Intergranular Cohesion in Iron", Solid State Comms., 1990, 76, 247-251
Cite error: <ref> tag with name "hooke" defined in <references> is not used in prior text.