Rep:Mod:littleliana2
HuiJun Liu's Computational labs Module 2: Week 1
Introduction
The goal of the first week of computational labs was to learn to use the program gaussview to observe different energetic and magnetic properties of desire molecules. Optimizations at different levels (basis sets) were carry out and checked for successes with a frequency analysis. Vibrations of desire molecules were looked at and a IR spectrum were generated by computational calculations. The use of gaussview in generating MO's and carrying out a NBO charge distribution analysis for desire molecules were also performed. For larger molecules that requires time consuming calculations, the departmental SCAN HPC service was explored and use to carry out such calculations. Many calculations were published on D-space to check for plagiarism.
Optimization
BH3
1st optimization (3-21G)
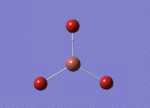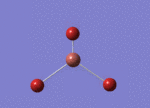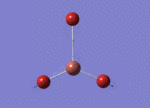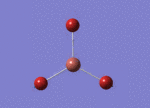
BH3 molecule created on Gaussview
All 3 B-H bond length changed to 1.5 A, it's angle and torsion angle was not changed.
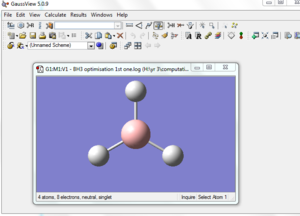
An optimization was run using:
- method: B3LYP
- basis set: 3-21G
The following results were obtained:
An log file of the optimization was obtained and can be viewed here
| File type | .log |
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Basis set | 3-21G |
| Charge | 0 |
| Spin | Singlet |
| Final Energy (a.u.) | -26.46226338 |
| Gradient (a.u.) | 0.00020672 |
| Dipole moment (D) | 0.0000 |
| Point Group | D3H |
| Job cpu time (s) | 29.0 |
The full results were viewed from the log file, and it was check that the optimization was actually completed by checking that the forces have been converged bellow the "items" table:
Item Value Threshold Converged?
Maximum Force 0.000413 0.000450 YES
RMS Force 0.000271 0.000300 YES
Maximum Displacement 0.001610 0.001800 YES
RMS Displacement 0.001054 0.001200 YES
Predicted change in Energy=-1.071764D-06
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1935 -DE/DX = 0.0004 !
! R2 R(1,3) 1.1935 -DE/DX = 0.0004 !
! R3 R(1,4) 1.1935 -DE/DX = 0.0004 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
The graphs for the total energy and the RMS gradient over the period of the optimization was open, and it was check again that the RMS gradient shows that the gradient for the optimized structure (at the end of the graphs, structure 4) was smaller then 0.001, thus it can be concluded that the optimization was completed.
Furthermore, it was noticed that there were no bonds present in the first two structures during the optimization of the gaussview BH3 molecule, it is understood that this is due to guassview's predefine criteria of atom distances in order for a bond to be drawn, not that there's no bond present.
The inquire button was used to determine:
- optimized B-H bond distance - 1.19349
- optimized H-B-H bond angle - 120.000
The total energy was found to be: -26.46226338 a.u.
2nd optimization (6-31G (d,p))
Further optimization of previous 3-21G optimized structure was done with a basis set of 6-31G(d,p), and the following results was obtained:
| File type | .log |
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Final Energy (a.u.) | -26.61532363 |
| Gradient (a.u.) | 0.00000235 |
| Dipole moment (D) | 0.0000 |
| Point Group | D3H |
| Job cpu time (s) | 7.0 |
The log file can be viewed here
An log file output of the molecule was obtained and it was checked that the forces did converged at the end of the optimization by checking the "items" table within the log file:
Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.000019 0.001800 YES
RMS Displacement 0.000012 0.001200 YES
Predicted change in Energy=-1.304899D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1923 -DE/DX = 0.0 !
! R2 R(1,3) 1.1923 -DE/DX = 0.0 !
! R3 R(1,4) 1.1923 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
The inquire button was used to determine the following information:
- optimized B-H bond distance - 1.19232
- optimized H-B-H bond angle - 120.000
The total energy was found to be: -26.61532363 a.u.
TlBr3
a TlBr3 molecule was created in gaussview and it's symmetry restricted to D3h with a very high tolerance of "very tight (0.0001)"
An optimization was run using:
- method: B3LYP
- basis set: LANL2DZ
An log file output of the molecule was obtained and can be viewed here
This file was submitted to D-space and sited as DOI:10042/21779
| File type | .log |
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Basis set | LANL2DZ |
| Charge | 0 |
| Spin | Singlet |
| Final Energy (a.u.) | -91.21812851 |
| Gradient (a.u.) | 0.00000090 |
| Dipole moment (D) | 0.0000 |
| Point Group | D3H |
| Job cpu time (s) | 10.0 |
"Items" table:
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000014 0.001200 YES
Predicted change in Energy=-6.084033D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.651 -DE/DX = 0.0 !
! R2 R(1,3) 2.651 -DE/DX = 0.0 !
! R3 R(1,4) 2.651 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
The inquire button was used to determine the following information:
- optimized Tl-Br bond distance - 2.65095
- optimized Br-Tl-Br bond angle - 120.000
The total energy was found to be: -91.21812851 a.u.
BBr3
From our 6-21G(d,p) optimized BH3 molecule, all three H atoms were changed to Br atoms.
An input file from gaussview was created using the following basis sets:
- 6-21G(d,p) for B atom
- LanL2DZ for Br atoms
The optimized log file was obtained and can be view here
| File type | .log |
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Basis set | Gen |
| Charge | 0 |
| Spin | Singlet |
| Final Energy (a.u.) | -64.43645296 |
| Gradient (a.u.) | 0.00000382 |
| Dipole moment (D) | 0.0000 |
| Point Group | D3H |
| Job cpu time (s) | 36.2 |
"items" table
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000036 0.001800 YES
RMS Displacement 0.000023 0.001200 YES
Predicted change in Energy=-4.026911D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.934 -DE/DX = 0.0 !
! R2 R(1,3) 1.934 -DE/DX = 0.0 !
! R3 R(1,4) 1.934 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
- optimized Tl-Br bond distance - 1.93396
- optimized Br-Tl-Br bond angle - 120.000
The total energy was found to be: -64.43645296 a.u.
This file was submitted to D-space and sited as DOI:10042/21786
Analysis of optimized bond lengths
| Compound | Bond distance (Å) |
|---|---|
| BH3 (3-21G) | 1.19349 |
| BH3 (6-31G) | 1.19232 |
| BBr3 | 1.93396 |
| TlBr3 | 2.65095 |
By changing the ligand from H to Br there were an obvious increase in bond length as expected, since Br is a far larger atom then the Hydrogen atom. Br has far more electrons, in s,p and d orbitals while as hydrogen only has 1 electron in the 1s orbital. Secondly, Br requires an electron to fill it's octet, while as hydrogen needs it only to fill it's s orbital. However, both atoms only require 1 more electron to fill its outer shell and thus, accepts electron density in a similar fashion and has relatively small outer shells.
By changing the central atom from B to Tl, an increase of bond length was observed again. This is due to the increase size of Tl as well as the fact that Tl now has much more diffuse valance orbitals. For interactions of such large orbitals, the orbital overlap is poor and thus weaker bonds are form compared to those smaller atoms with smaller orbitals. Both Tl and B are in group 3 and thus, have the same number of valence electrons. However, Tl is a far larger atom then B and is much more down the group, thus it is observed that Tl can undergo the inert pair effect where it forms a lower oxidations state due to the increase energy gap between it's valence s and p orbitals in comparison to B.
A bond is the electrons shared between two atoms in an molecular orbital that was form from interactions between the two atomic orbitals of the relavent atoms. These interactions are distance dependent and weakens as the two atoms are further appart. Gaussview does not draw in the bonds between atoms when they are above an certain distance, but this does not mean that there are no bond present, there is still an interaction, just weaker.
Vibrational / Frequency Analysis
BH3
From a copy of our 6-31G(d,p) optimized BH3 log file a frequency calculation was ran on gaussview and the following results were found:
| File type | .log |
| Calculation type | FREQ |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Final Energy (a.u.) | -26.61532363 |
| Gradient (a.u.) | 0.00000237 |
| Dipole moment (D) | 0.0000 |
| Point Group | D3H |
| Job cpu time (s) | 5.0 |
The log file calculation can be viewed here
The calculation was shown to be ok by the values of the "low frequencies" being smaller then 15cm-1
Full mass-weighted force constant matrix:
Low frequencies --- -0.9033 -0.7343 -0.0054 6.7375 12.2491 12.2824
Low frequencies --- 1163.0003 1213.1853 1213.1880
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A2" E' E'
Frequencies -- 1163.0003 1213.1853 1213.1880
Red. masses -- 1.2531 1.1072 1.1072
Frc consts -- 0.9986 0.9601 0.9601
IR Inten -- 92.5478 14.0553 14.0589
Atom AN X Y Z X Y Z X Y Z
1 5 0.00 0.00 0.16 0.00 0.10 0.00 -0.10 0.00 0.00
2 1 0.00 0.00 -0.57 0.00 0.08 0.00 0.81 0.00 0.00
3 1 0.00 0.00 -0.57 -0.39 -0.59 0.00 0.14 0.39 0.00
4 1 0.00 0.00 -0.57 0.39 -0.59 0.00 0.14 -0.39 0.00
4 5 6
A1' E' E'
Frequencies -- 2582.2624 2715.4311 2715.4323
Red. masses -- 1.0078 1.1273 1.1273
Frc consts -- 3.9595 4.8976 4.8976
IR Inten -- 0.0000 126.3307 126.3211
 |
There are only three visible peaks on this IR graphs even though there was obviously 6 different vibrations present for the BH3 molecule. Vibration number 4, as stated in the table above, is perfectly symmetrical and does not induce an dipole in the molecule, thus is not expected to be IR active. This leaves five peaks that we expected to see on our graph. The data obtain from gaussview has shown that vibration 2 and 3 both occur at frequency of 1213.19 cm-1 with the exact same intensity, thus it's no wonder that only one peak is observed for these two bends. Vibrations 5 and 6 also occur at similar frequencies and intensities, careful analysis of the graph show that there are indeed two peaks at around 2700cm-1, thus our graph did agree with gaussview data in the number of peaks present.
TlBr3
An frequency analysis was ran on the optimized TlBr3 molecule using the SCAN HPC service and the results publish on D-space: DOI:10042/21969
The full log file can be viewed here
| File type | .log |
| Calculation type | FREQ |
| Calculation method | RB3LYP |
| Basis set | LANL2DZ |
| Charge | 0 |
| Spin | Singlet |
| Final Energy (a.u.) | -91.21812851 |
| Gradient (a.u.) | 0.00000088 |
| Dipole moment (D) | 0.0000 |
| Point Group | D3H |
| Job cpu time (s) | 28.8 |
Low frequencies --- -3.4213 -0.0026 -0.0004 0.0015 3.9367 3.9367
Low frequencies --- 46.4289 46.4292 52.1449
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
E' E' A2"
Frequencies -- 46.4289 46.4292 52.1449
Red. masses -- 88.4613 88.4613 117.7209
Frc consts -- 0.1124 0.1124 0.1886
IR Inten -- 3.6867 3.6867 5.8466
 |
Comparison of TlBr3 and BH3 frequencies
| No. | BH3 (cm-1) | TlBr3(cm-1) |
| 1 | 1163.00 | 46.43 |
| 2 | 1213.19 | 46.43 |
| 3 | 1213.19 | 52.14 |
| 4 | 2582.26 | 165.27 |
| 5 | 2715.43 | 210.69 |
| 6 | 2715.43 | 210.69 |
The large difference in frequencies between the two compounds indicate that there are a large difference in the energy of the B-H bond compare to that of the Tl-Br bond, as expected. The Tl-Br bonds are a lot weaker than that of the BH bonds, thus they do not require as much energy to bend it's bonds. However, since TlBr3 is a much larger molecule then BH3, it will vibrate much less then that of the small BH3, Bends would also be much more stericly hindered.
There were an reordering in modes, where the A2" mode had an lower energy then E' in the BH3 molecule while in the TlBr3, the A2" had an higher energy then E'. This can be account for by the fact that Tl is a much larger center then the B atom, so when moving in the A2" mode, there is a much greater steric hindrance and thus, requires more energy then it would when moving in the E, while as for the BH3 molecule, since B is so small, steric hindrance is minmal and so E requires more energy instead.
The two spectra show the same number of vibrations for the two molecules as expected. Their overall shape is quite similar and both spectra had the same modes, if not at the same relative energies. Both had a vibration of zero frequency, as no dipole was formed, and both spectra had two pairs of vibrations at almost identical frequencies. The vibrations were two bends and two stretches, where the A2" and E' bend will require similar energy, they would need a lot less then a stretch, thus the A1' and E' stretch vibrations lies close together but far from the two bend modes.
The same method and basis set for both the optimisation and frequency is required since the purpose of carrying out the frequency analysis is to check that our optimization is a minimum and thus, completed, if we used a different basis set for the frequency analysis, it would be to determine the optimization minimum for a different optimization, and thus, unable to serve our purpose.
Another purpose of doing the frequency analysis is to predict the gas phase IR spectra, so that actual experimental lab work can be avoided, since some molecules, such as Tl, are highly toxic and would be best if we can not have to handle it.
The low frequencies are the motion of the center of mass of the molecule, and can be expected to be much smaller then our actual vibrational modes.
Molecular Orbitals of BH3
Calculations done on SCAN HPC computers and submitted to D-space: DOI:10042/21923
The full formatted checkpoint file can be viewed here

|
| Symmetry Label | LCAO MO | Calculated MO |
| a1' |  |

|
| e' | 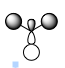 |

|
| e' | 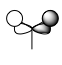 |

|
| a2' | 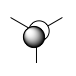 |

|
As expected, my calculated MO is highly similar to that of the LCAO theory. This tells us that LCAO is quite a reliable theory in that their predicted MO's are quite similar to that we find in reality.
NH3
Optimization and Analysis
6-31(d,p) done using gaussview
full log file can be viewed here
| File type | .log |
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Final Energy (a.u.) | -56.55776856 |
| Gradient (a.u.) | 0.00000885 |
| Dipole moment (D) | 1.8464 |
| Point Group | C1 |
| Job cpu time | 11 min 46.0 sec |
Item Value Threshold Converged?
Maximum Force 0.000024 0.000450 YES
RMS Force 0.000012 0.000300 YES
Maximum Displacement 0.000079 0.001800 YES
RMS Displacement 0.000053 0.001200 YES
Predicted change in Energy=-1.629727D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7413 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7486 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7479 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8631 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Frequency Analysis
full log file can be viewed here
| File type | .log |
| Calculation type | FREQ |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Final Energy (a.u.) | -56.55776856 |
| Gradient (a.u.) | 0.00000878 |
| Dipole moment (D) | 1.8464 |
| Point Group | C1 |
| Job cpu time | 4 min 33.0 sec |
Full mass-weighted force constant matrix:
Low frequencies --- -30.8045 -0.0017 -0.0013 -0.0012 20.2188 28.2150
Low frequencies --- 1089.5530 1694.1235 1694.1861
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A A
Frequencies -- 1089.5530 1694.1235 1694.1861
Red. masses -- 1.1800 1.0644 1.0644
Frc consts -- 0.8253 1.8000 1.8001
IR Inten -- 145.4405 13.5558 13.5561
Atom AN X Y Z X Y Z X Y Z
1 7 0.12 0.00 0.00 0.00 -0.02 -0.06 0.00 0.06 -0.02
2 1 -0.53 -0.21 0.00 -0.07 -0.04 0.73 0.25 0.14 0.19
3 1 -0.53 0.11 0.18 0.25 -0.24 -0.02 -0.07 -0.61 0.40
4 1 -0.53 0.11 -0.18 -0.18 0.52 0.18 -0.18 -0.41 -0.36
4 5 6
A A A
Frequencies -- 3460.9836 3589.4000 3589.5263
Red. masses -- 1.0272 1.0883 1.0883
Frc consts -- 7.2496 8.2615 8.2621
IR Inten -- 1.0593 0.2700 0.2708
no negative frequencies were observed:
| Vibration | Frequency | Infrared |
|---|---|---|
| 1 | 1089.55 | 145.4405 |
| 2 | 1694.12 | 13.5558 |
| 3 | 1694.19 | 13.5561 |
| 4 | 3460.98 | 1.0593 |
| 5 | 3589.40 | 0.2700 |
| 6 | 3589.53 | 0.2708 |
Population analysis
submitted to D-space: DOI:10042/21946
full formatted checkpoint file can be viewed here
| File type | .fch |
| Calculation type | sp |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Final Energy (a.u.) | -56.55776856 |
| Gradient (a.u.) | 0.00000878 |
| Dipole moment (D) | 1.8464 |
| Point Group | C1 |
| Job cpu time | 2 min 33.0 sec |
NBO analysis
The full log file from population analysis can be viewed Media:Log_NH3_NBO_Huijun_Liu.log
This was published on D-space: DOI:10042/21946
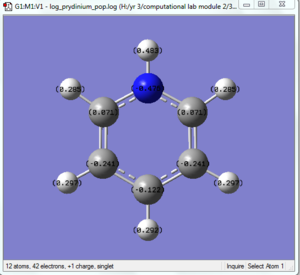
The color range for the NBO was +1.000 to -1.000 for the colors bright red to bright green respectively. The specific NBO charge for the nitrogen atom was -1.125 and the charge for the hydrogen atoms were 0.375 each.

|
NH3BH3
Optimization
Due to the large number of atoms in the NH3BH3 molecule, the optimization for it was done via two steps starting first with a 3-21G basis set and then moving on to 6-31G(d,p)
1st Optimization: 3-21G
The full log file of the optimization was obtained and can be viewed here
| File type | .log |
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Basis set | 3-21G |
| Charge | 0 |
| Spin | Singlet |
| Final Energy (a.u.) | -82.76661837 |
| Gradient (a.u.) | 0.00003006 |
| Dipole moment (D) | 5.8431 |
| Point Group | C1 |
| Job cpu time (s) | 31.0 |
It was check that the forces have converged in the "items" table
Item Value Threshold Converged?
Maximum Force 0.000094 0.000450 YES
RMS Force 0.000030 0.000300 YES
Maximum Displacement 0.000419 0.001800 YES
RMS Displacement 0.000178 0.001200 YES
Predicted change in Energy=-5.742833D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,7) 1.0277 -DE/DX = -0.0001 !
! R2 R(2,7) 1.0277 -DE/DX = -0.0001 !
! R3 R(3,7) 1.0277 -DE/DX = 0.0 !
! R4 R(4,8) 1.212 -DE/DX = 0.0 !
! R5 R(5,8) 1.212 -DE/DX = -0.0001 !
! R6 R(6,8) 1.212 -DE/DX = -0.0001 !
! R7 R(7,8) 1.6854 -DE/DX = -0.0001 !
! A1 A(1,7,2) 109.3469 -DE/DX = 0.0 !
! A2 A(1,7,3) 109.35 -DE/DX = 0.0 !
! A3 A(1,7,8) 109.5925 -DE/DX = 0.0 !
! A4 A(2,7,3) 109.3462 -DE/DX = 0.0 !
! A5 A(2,7,8) 109.5973 -DE/DX = 0.0 !
! A6 A(3,7,8) 109.5935 -DE/DX = 0.0 !
! A7 A(4,8,5) 113.563 -DE/DX = 0.0 !
! A8 A(4,8,6) 113.5594 -DE/DX = 0.0 !
! A9 A(4,8,7) 104.9872 -DE/DX = 0.0 !
! A10 A(5,8,6) 113.5575 -DE/DX = 0.0 !
! A11 A(5,8,7) 104.9846 -DE/DX = 0.0 !
! A12 A(6,8,7) 104.9848 -DE/DX = 0.0 !
! D1 D(1,7,8,4) 179.9938 -DE/DX = 0.0 !
! D2 D(1,7,8,5) -60.0024 -DE/DX = 0.0 !
! D3 D(1,7,8,6) 59.994 -DE/DX = 0.0 !
! D4 D(2,7,8,4) -60.0065 -DE/DX = 0.0 !
! D5 D(2,7,8,5) 59.9972 -DE/DX = 0.0 !
! D6 D(2,7,8,6) 179.9936 -DE/DX = 0.0 !
! D7 D(3,7,8,4) 59.9928 -DE/DX = 0.0 !
! D8 D(3,7,8,5) 179.9966 -DE/DX = 0.0 !
! D9 D(3,7,8,6) -60.007 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
2nd Optimization: 6-31G(d,p)
The full log file of the optimization was obtained and can be viewed here
| File type | .log |
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Final Energy (a.u.) | -83.22469010 |
| Gradient (a.u.) | 0.00005646 |
| Dipole moment (D) | 5.5623 |
| Point Group | C1 |
| Job cpu time (s) | 31.0 |
It was check that the forces have converged in the "items" table
Item Value Threshold Converged?
Maximum Force 0.000132 0.000450 YES
RMS Force 0.000037 0.000300 YES
Maximum Displacement 0.001194 0.001800 YES
RMS Displacement 0.000532 0.001200 YES
Predicted change in Energy=-1.177062D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,7) 1.0185 -DE/DX = 0.0 !
! R2 R(2,7) 1.0185 -DE/DX = 0.0 !
! R3 R(3,7) 1.0185 -DE/DX = 0.0 !
! R4 R(4,8) 1.2097 -DE/DX = 0.0 !
! R5 R(5,8) 1.2097 -DE/DX = 0.0 !
! R6 R(6,8) 1.2097 -DE/DX = 0.0 !
! R7 R(7,8) 1.6686 -DE/DX = -0.0001 !
! A1 A(1,7,2) 107.8674 -DE/DX = 0.0 !
! A2 A(1,7,3) 107.8625 -DE/DX = 0.0 !
! A3 A(1,7,8) 111.0305 -DE/DX = 0.0 !
! A4 A(2,7,3) 107.8643 -DE/DX = 0.0 !
! A5 A(2,7,8) 111.0345 -DE/DX = 0.0 !
! A6 A(3,7,8) 111.0356 -DE/DX = 0.0 !
! A7 A(4,8,5) 113.901 -DE/DX = 0.0 !
! A8 A(4,8,6) 113.906 -DE/DX = 0.0 !
! A9 A(4,8,7) 104.5592 -DE/DX = 0.0001 !
! A10 A(5,8,6) 113.9 -DE/DX = 0.0 !
! A11 A(5,8,7) 104.5627 -DE/DX = 0.0 !
! A12 A(6,8,7) 104.5635 -DE/DX = 0.0 !
! D1 D(1,7,8,4) 180.0372 -DE/DX = 0.0 !
! D2 D(1,7,8,5) -59.965 -DE/DX = 0.0 !
! D3 D(1,7,8,6) 60.0335 -DE/DX = 0.0 !
! D4 D(2,7,8,4) -59.9607 -DE/DX = 0.0 !
! D5 D(2,7,8,5) 60.037 -DE/DX = 0.0 !
! D6 D(2,7,8,6) 180.0355 -DE/DX = 0.0 !
! D7 D(3,7,8,4) 60.0407 -DE/DX = 0.0 !
! D8 D(3,7,8,5) 180.0384 -DE/DX = 0.0 !
! D9 D(3,7,8,6) -59.9631 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Frequency Analysis
full log file can be viewed here
| File type | .log |
| Calculation type | FREQ |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Final Energy (a.u.) | -83.22469011 |
| Gradient (a.u.) | 0.00005640 |
| Dipole moment (D) | 5.5623 |
| Point Group | C1 |
| Job cpu time | 38.0 sec |
It was check that the low frequencies was reasonable and a minimum was observed, proving that the optimization was completed.
Low frequencies --- -0.0011 0.0006 0.0012 2.1062 15.9915 20.4431
Low frequencies --- 263.3672 631.3120 638.1575
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A A
Frequencies -- 263.3627 631.3120 638.1574
Red. masses -- 1.0078 5.0011 1.0452
Frc consts -- 0.0412 1.1744 0.2508
IR Inten -- 0.0000 14.1187 3.5703
Atom AN X Y Z X Y Z X Y Z
1 1 0.00 -0.24 0.38 0.35 0.00 0.00 -0.12 0.14 -0.16
2 1 0.00 -0.21 -0.40 0.35 0.00 0.00 0.56 0.12 -0.14
3 1 0.00 0.45 0.02 0.37 0.00 0.01 -0.44 0.15 -0.12
4 1 0.00 -0.19 0.31 -0.29 -0.03 -0.01 -0.09 0.10 -0.12
5 1 0.00 0.36 0.01 -0.28 0.00 0.04 -0.34 0.11 -0.08
6 1 0.00 -0.17 -0.32 -0.29 0.03 -0.01 0.43 0.07 -0.09
7 7 0.00 0.00 0.00 0.36 0.00 0.00 0.00 -0.03 0.03
8 5 0.00 0.00 0.00 -0.48 0.00 0.00 0.00 -0.02 0.02
Association Energy Calculations
Taking all our results from the 6-31G(d,p) level optimization, the following energies were obtained
- E(NH3)= -56.55776856 a.u
- E(BH3)= -26.61532363 a.u
- E(NH3BH3)= -83.22469011 a.u
It can be reasoned that the association energy is just the difference between the product and the combined reactants, thus:
AC Energy= ΔE =E(NH3BH3)-[E(NH3)+E(BH3)]
= -83.22469011+56.55776856+26.61532363 = -0.05159792 a.u
to convert a.u units to kJ/mol the following equation was used:
1 kJ/mol= 2625.5 a.u
Thus our calculated association energy is: -135.47 kJ/mol
This number is quite reasonable for a association energy of a reaction, so can be consider correct.
Note on Accuracy: energy has an error of ~10 kJ/mol, which is 0.00380879 a.u, though we been told to record our energies to 8 decimal places in a.u units.
HuiJun Liu's Computational labs Module 2: Week 2
Benzene
6-31G(d,p) Optimization
Calculations done on SCAN HPC computers and submitted to D-space: DOI:10042/21939
The full log file of the optimization was obtained and can be viewed here
| File type | .log |
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Final Energy (a.u.) | -232.25820551 |
| Gradient (a.u.) | 0.00009549 |
| Dipole moment (D) | 0.0001 |
| Point Group | C1 |
| Job cpu time | 2 min 2.9 sec |
The "items" table was check to prove that the forces have indeed converged:
Item Value Threshold Converged? Maximum Force 0.000212 0.000450 YES RMS Force 0.000085 0.000300 YES Maximum Displacement 0.000991 0.001800 YES RMS Displacement 0.000315 0.001200 YES Predicted change in Energy=-5.157454D-07 Optimization completed. -- Stationary point found.
Frequency Analysis
Calculations done on SCAN HPC computers and submitted to D-space: DOI:10042/21944
The full log file was obtained and can be viewed here
| File type | .log |
| Calculation type | FREQ |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Final Energy (a.u.) | -232.25820551 |
| Gradient (a.u.) | 0.00009549 |
| Dipole moment (D) | 0.0001 |
| Point Group | C1 |
| Job cpu time | 5 min 31.0 sec |
The low frequencies were checked to see that they were reasonable and no negative frequency were observed.
Low frequencies --- -17.2788 -14.5868 -9.6527 -0.0008 -0.0006 -0.0005
Low frequencies --- 413.7971 414.4697 620.8545
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A A
Frequencies -- 413.7971 414.4697 620.8540
Red. masses -- 2.9404 2.9434 6.0705
Frc consts -- 0.2966 0.2979 1.3786
IR Inten -- 0.0000 0.0000 0.0000
Population Analysis
Calculations done on SCAN HPC computers and submitted to D-space: DOI:10042/22032
The full log file was obtained and can be viewed here
The full fchk file can be viewed here
| File type | .log |
| Calculation type | SP |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Final Energy (a.u.) | -232.25820551 |
| Gradient (a.u.) | 0.00009549 |
| Dipole moment (D) | 0.0001 |
| Point Group | C2V |
| Job cpu time | 5 min 31.0 sec |
| Sigma All bonding | Pi all bonding | pi HOMO |
 |
 |
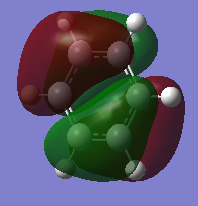 |
 |
 |
Boratabenzene
6-31G(d,p) Optimization
Calculations done on SCAN HPC computers and submitted to D-space: DOI:10042/21940
The full log file of the optimization was obtained and can be viewed here
| File type | .log |
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Charge | -1 |
| Spin | Singlet |
| Final Energy (a.u.) | -219.02052984 |
| Gradient (a.u.) | 0.00015840 |
| Dipole moment (D) | 2.8465 |
| Point Group | C1 |
| Job cpu time | 3 min 59.0 sec |
The "items" table was check to prove that the forces have indeed converged:
Item Value Threshold Converged? Maximum Force 0.000159 0.000450 YES RMS Force 0.000069 0.000300 YES Maximum Displacement 0.000878 0.001800 YES RMS Displacement 0.000326 0.001200 YES Predicted change in Energy=-6.589451D-07 Optimization completed. -- Stationary point found.
Frequency Analysis
Calculations done on SCAN HPC computers and submitted to D-space: DOI:10042/22020
The full log file was obtained and can be viewed here
| File type | .log |
| Calculation type | FREQ |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Charge | -1 |
| Spin | Singlet |
| Final Energy (a.u.) | -219.02052984 |
| Gradient (a.u.) | 0.00015838 |
| Dipole moment (D) | 2.8465 |
| Point Group | C1 |
| Job cpu time | 5 min 34.0 sec |
It was checked that the low frequencies were reasonable and that there were no negative frequencies present:
Low frequencies --- -13.1275 -0.0007 -0.0005 0.0002 15.0447 18.1653
Low frequencies --- 371.3454 404.2334 565.2534
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A A
Frequencies -- 371.3453 404.2327 565.2534
Red. masses -- 2.6885 3.2177 5.7648
Frc consts -- 0.2184 0.3098 1.0852
IR Inten -- 2.3000 0.0000 0.1576
Population Analysis
Calculations done on SCAN HPC computers and submitted to D-space: DOI:10042/22049
The full log file was obtained and can be viewed here
The full fchk was obtained and can be viewed here
| File type | .log |
| Calculation type | SP |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Charge | -1 |
| Spin | Singlet |
| Final Energy (a.u.) | -219.02052984 |
| Gradient (a.u.) | 0.0000000 |
| Dipole moment (D) | 2.8466 |
| Point Group | C1 |
| Job cpu time | 5 min 31.0 sec |
| Sigma All bonding | Pi all bonding | pi HOMO |
 |
 |
 |
 |
 |
B-H Bond particularly long
Pyridinium
6-31G(d,p) Optimization
Calculations done on SCAN HPC computers and submitted to D-space: DOI:10042/21941
The full log file of the optimization was obtained and can be viewed here
| File type | .log |
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Charge | 1 |
| Spin | Singlet |
| Final Energy (a.u.) | -248.66807396 |
| Gradient (a.u.) | 0.00003894 |
| Dipole moment (D) | 1.8727 |
| Point Group | C1 |
| Job cpu time | 4 min 0.3 sec |
The "items" table was check to prove that the forces have indeed converged:
Item Value Threshold Converged? Maximum Force 0.000065 0.000450 YES RMS Force 0.000023 0.000300 YES Maximum Displacement 0.000826 0.001800 YES RMS Displacement 0.000176 0.001200 YES Predicted change in Energy=-6.972574D-08 Optimization completed. -- Stationary point found.
Frequency Analysis
Calculations done on SCAN HPC computers and submitted to D-space: DOI:10042/22021
The full log file was obtained and can be viewed here
| File type | .log |
| Calculation type | FREQ |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Charge | 1 |
| Spin | Singlet |
| Final Energy (a.u.) | -248.66807396 |
| Gradient (a.u.) | 0.00003897 |
| Dipole moment (D) | 1.8727 |
| Point Group | C1 |
| Job cpu time | 5 min 39.5 sec |
It was checked that the low frequencies were reasonable and that there were no negative frequencies present:
Full mass-weighted force constant matrix:
Low frequencies --- -7.2115 -0.0008 -0.0006 -0.0004 17.3444 18.5499
Low frequencies --- 392.4572 404.0614 620.4717
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A A
Frequencies -- 392.4571 404.0614 620.4717
Red. masses -- 2.9474 2.7453 6.2544
Frc consts -- 0.2675 0.2641 1.4187
IR Inten -- 0.9649 0.0000 0.0144
Population Analysis
Calculations done on SCAN HPC computers and submitted to D-space: DOI:10042/22053
The full log file was obtained and can be viewed here
The full fchk file was obtained and can be viewed here
| File type | .log |
| Calculation type | SP |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Charge | 1 |
| Spin | Singlet |
| Final Energy (a.u.) | -248.66807396 |
| Gradient (a.u.) | 0.00009549 |
| Dipole moment (D) | 1.8727 |
| Point Group | C1 |
| Job cpu time | 5 min 31.0 sec |
| Sigma All bonding | Pi all bonding | pi HOMO |
 |
 |
 |
 |
 |
Borazine
6-31G(d,p) Optimization
Calculations done on SCAN HPC computers and submitted to D-space: DOI:10042/21942
The full log file of the optimization was obtained and can be viewed here
| File type | .log |
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Final Energy (a.u.) | -242.68459789 |
| Gradient (a.u.) | 0.00007128 |
| Dipole moment (D) | 0.0003 |
| Point Group | C1 |
| Job cpu time | 5 min 11.6 sec |
The "items" table was check to prove that the forces have indeed converged:
Item Value Threshold Converged? Maximum Force 0.000117 0.000450 YES RMS Force 0.000036 0.000300 YES Maximum Displacement 0.000327 0.001800 YES RMS Displacement 0.000104 0.001200 YES Predicted change in Energy=-1.206131D-07 Optimization completed. -- Stationary point found.
Frequency Analysis
Calculations done on SCAN HPC computers and submitted to D-space: DOI:10042/22022
The full log file was obtained and can be viewed here
| File type | .log |
| Calculation type | FREQ |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Final Energy (a.u.) | -242.68459788 |
| Gradient (a.u.) | 0.00007127 |
| Dipole moment (D) | 0.0003 |
| Point Group | C1 |
| Job cpu time | 5 min 31.6 sec |
It was checked that the low frequencies were reasonable and that there were no negative frequencies present:
Full mass-weighted force constant matrix:
Low frequencies --- -11.1593 -0.0004 0.0003 0.0007 8.7441 11.1735
Low frequencies --- 288.5026 290.4077 404.0135
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A A
Frequencies -- 288.5025 290.4075 404.0130
Red. masses -- 2.9264 2.9236 1.9244
Frc consts -- 0.1435 0.1453 0.1851
IR Inten -- 0.0000 0.0000 23.6788
Population Analysis
Calculations done on SCAN HPC computers and submitted to D-space: DOI:10042/22054
The full log file was obtained and can be viewed here
The full fchk file was obtained and can be viewed here
| File type | .log |
| Calculation type | SP |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Final Energy (a.u.) | -242.68459789 |
| Gradient (a.u.) | 0.00009549 |
| Dipole moment (D) | 0.0001 |
| Point Group | C1 |
| Job cpu time | 4 min 31.0 sec |
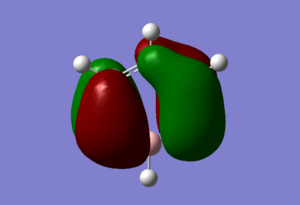
| Sigma All bonding | Pi all bonding | pi HOMO |
 |
 |
 |
 |
 |
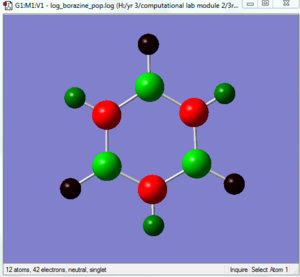
B-H bond shorter, B-N bonds longer
Discussion and Analysis
Charge Distribution Analysis
Our reference benzene molecule displayed a highly even distribution of it's electron charge. All six carbon on the benzene had a -0.239 charge and all six hydrogen atoms had a 0.239 positive charge. This is as expected, since all six carbon atom of the benzene molecule has the same size and energy AO's that are involved in the splitting to make benzene's MO's, thus all six carbon atoms are in the same environment and does not differ from each another. The difference between electronegativity of the carbon atom and hydrogen atom give raise to the majority of the electron cloud to form around the carbon atom, thus giving a slight dipole on the C-H bond, where carbon is slight electronegative and hydrogen, positive. Since benzene is a highly symmetrical molecule, these individual C-H bond dipoles cancel each other out to give an overall molecule of no dipole. It's also noted that the exact charge numbers of the two atoms are exactly the same, and the sum of all the charges on all the atoms gives us an overall zero charge for the molecule, as expected.
The boratabenzene, on the other hand, not only displayed a difference in charge between the boran atom and the carbon atoms as would be expected due to their difference in electronegativity, but also display an difference in electron charge between the carbon atoms themselves. The two carbon atoms directly next to the boran atoms have the most negative charge (-0.588), the two carbon atoms that are 2 bond lengths away have the smallest negative charge of all the carbon atoms (-0.250), and the carbon atom that is furthest away from the the boran atom has the second most negative charge of the molecule (-0.340). This difference in due to the relative electropositive boran atom in comparison to the carbon atoms, so the two carbon atoms next to the borans experience less pull from the boran atom then the pull it gets from the carbon atom next to it, thus the carbon atoms next to the boran holds on to it's electrons density better. A more technical view can be seen by the fact that boran has higher energy LCAO's then those of the carbon LCAO, thus the MO's that are made from the combination of these LCAO's will display more carbon AO characteristic then boran characteristic since they are closer in energy. The relative high numbers for these charges are due to the fact that the overall molecule has a -1 charge. When the boratabenzene molecule was created in gaussview, a negative charge was added to make this molecule isoelectronic to that of the benzene molecule, this negative charge is formally drawn on the boran atom, but our NBO analysis here showed that the boran actually has a slightly positive charge (0.202) and the negative charge had been displace onto the five carbon atoms instead. The hydrogen's also have differing charges from each other, with the hydrogen on the boran atom being the only negatively charged hydrogen (-0.095) due to the formal negative charge on the boran atom. All the other hydrogen atoms show the same trend in their charges as their carbon counterparts due to the charge on their carbon.
The prydinium molecule shows a similar trend to that of the boratabenzene molecule. The prydinium molecule has an overall charge of +1 which was formally place on the nitrogen atom to insure it's isoelectric to benzene. However, our NBO analysis show that the nitrogen in reality has a -0.470 negative charge due to it's relative high electronegativity in comparison to the carbon and hydrogen atoms in the ring, Thus, the positive charge was disperse between the Hydrogen on the nitrogen atom (0.483, highest positive charge of all the atoms) and the 5 carbons on the ring. The two carbons next to the nitrogen showed a positive charge of 0.071 and the carbon atom furthest away from the nitrogen had a -0.122 charge, both significantly higher than that of the reference benzene carbons, showing significant affect of the nitrogen positive charge on it. The reasons are the same as stated for the boratabenzene, but only that due to the higher electronegativity of the nitrogen, it's LCAO's now lies lower in energy instead of higher, thus the bonding MO's will show more nitrogen characteristic then carbon. The other hydrogen atoms also follows the trend of the boratabenzene and are different from those of the benzene molecule. There is a observed dipole for both boratabenzene and prydinium as expected from their charge, this makes them much more reactive than that of benzene. Furthermore, since their are an obvious different between charge of the carbon atoms, chemical reactions can be tuned to get regiospecific attack of desire carbon positions (meta vs ortho, etc) while as with benzene alone, this can not be achieved.
The Borazine molecule no doubt has characteristic that are the most similar to that of the benzene molecule. The NBO shows that there are a negative charge on the nitrogen (-1.182) and a positive charge on boran (0,747). This is due to the highly polarize N-B bond where nitrogen is strongly electronegative and thus, draws the electron density away from the more electropositive boran. The hydrogen atoms had charges of (0.432) for those bonded to nitrogen atoms and (-0.077) for those bonded to boran atoms. This can be explained simply by the relative electronagtivities of nitrogen and boran. For example, for the N-H bond, since nitrogen is much more electronegative, it is expected that the nitrogen will have a negative charge while the hydrogen has a positive one. It is noted that due to the high level of symmetry of the molecule, each dipole moment and charge cancels each other out to leave us with a overall neutral molecule of zero dipole. However, since the ring atoms, nitrogen and boran are much more highly charged than the carbon atoms on the benzene molecule, it can be expected that the borazine molecule will be a lot more reactive then that of benzene.
Molecular orbitals analysis
The sigma totally bonding orbital, pi totally bonding orbita, and pi highest occupied molecular orbital (HOMO) were chosen for analysis, as seen above. It was noted that the cord orbital have far lower energies then the valence MO's and do not interact with each other and are thus, irrelevant for analysis. The generated MO all agreed with what would have been expected due to relative electronegativity of the different atoms, and the trends for the three analogs in comparison to our benzene "reference" were consistant for all three MO studied.
For the sigma totally bonding orbital (orbital 7 on gauss view) consist of six sigma bonds all in line and has the lowest energy of all the valence orbitals for all four complex. Relative to Benzene, where the computed MO showed a star shape of all the electron density spreading out evenly over the entire molecule, boratabenzene displayed a absence of electron density around the substituted boran atom while prydinium, on the other hand, had an MO where there were an obviously higher electron density around the Nitrogen. These observations are due to the fact that nitrogen has a higher electronegativity relative to the other carbon atoms, (while boran has a lower electronegativity) and thus are more capable of drawing electrons toward it, thus shifting the electron density toward the nitrogen atom and thus, away from the carbon atoms furthest away. Boran on the boratabenzene had the opposite effect, leaving the carbon atoms furthest away from the boran atom to have more electron density then those closer. The borazine molecule generated a MO of a triangular shape, where a high proportion of the electron density is centered around the three nitrogen atoms and away from the boran atoms.
The energy for the boratabenzene analog was higher (thus less negative) that that of the benzene molecule, while the prydinium molecule has a lower energy respectively. The MO for Borazine was very close in energy to that of benzene, only very slightly smaller. These are due to fact that the nitrogen has AO's lower in energy than that of the carbon atom AO's due to it's higher electronegativity, thus a lowering in the MO energy is observed. The same applies for the boratabenzene analog, where a raising in MO energy is observed. The borazine molecule has three borans which raise the MO's energy in comparison to benzene, and 3 nitrogens that lowers the energy in comparison, the overall effect was a average of the two effects that yield an energy very similar to that of the benzene molecule, with the nitrogen atoms slightly stronger in lowering the MO's then the borans are in raising it.
There were no degenerate orbital for this MO for all four molecules.
Orbital 17, the pi totally bonding orbital are six pi bonding orbitals from 6 p orbitals all in the same direction, and are highly symmetrical. The molecular orbitals themselves takes the form as expected with two electron clouds of opposite charge above and below the plane of the ring. Similar trends was observed as with the sigma orbital above, where there were more electron density around the nitrogen atom and less around the boran atom for prydinium and boratabenzene respectively. The borazine displayed two lobes of triangular shape electron clouds for reasons as discussed for orbital 7.
The energy for this MO also showed the same trend as seen for the sigma totally bonding MO, which boratabenzene having a higher energy, prydinium with a lower energy, and borazine a very slight lower energy in comparison to benzene. No degenerate MOs were found for this energy level as well.
The pi highest occupied molecular orbital (HOMO) are form from 6 p orbital of which three points to one direction and are next to each other, and three points in the other and are next to each other. The overall effect is a node through the middle of the molecule and four lobes of electron density. While benzene is highly symmetrical and display similar sizes for all the electron clouds, the analogs continue to display the same characteristics discuss above for the sigma MO, where there are less electron density around the boran atom for boratabenzene and more around the nitrogen for prydinium. The same energy trends were obtained as well.
The major difference for this MO is that there were an degenerate pi MO for benzene and borazine, but not for boratabenzene and prydinium. This is due to the fact that the insertion of only one boran of nitrogen atom change the symmetry of the Benzene molecule from the highly symmetrical D6h to much lower group. This causes the energy levels to vary. This observation can also be seen by the electron clouds of the MO, where with benzene, since all the electron density are spread out evenly, the two degenerate MO looks highly similar, but once there is a boran of nitrogen atom there to change this distribution, a clear difference was observed.
Over all, the effect of substitution of an carbon atom with an Nitrogen lowers one set of LCAO's that contributes to the MO, thus all the energy levels that interact with this set of LCAO, namely all except two sigma bonds and all of the pi bonds will be stabilized due to the relative higher elctronegativity of the nitrogen atom, thus an lowering of these MO's would be observed. The same goes for boran except that boran will destabilized and thus raise the energy of the MO's.