Rep:Mod:lh106module2
BH3 Introduction Module
A molecule of BH3 was created and optimised using Gaussian with the B3LYP method and a 3-21G base set (so quite a rough approximation to start with)to give the optimised molecule. The B3LYP method is a combinatin of the Lee, Yang and Parr correction that was created to fit the helium atom and B3 hybrid exchange function. It is therefore definitely an approximation as it only accurately fits helium.[1]
BCl |
| Optimised B-H bond distance | 1.5A |
| Optimised H-B-H angle | 120° |
| File type | .log |
| Calculation type | SP |
| Calculation method | RB3LYP |
| Base set | 3-21G |
| Final energy | -26.38297143au |
| Dipole moment | 0 Debye |
| Point group | D3H |
| Length of time | 10 seconds |
From this optimised molecule, the overall energy could be observed:
Vibrational Modes
The vibrational modes of BH3 were also calculated using Gaussian. Gaussian was also used to visualise the vibrational modes

| No. | Form of Vibration | Frequency | Intensity | Symmetry
D3H point group |
| 1 | 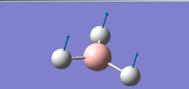
|
1145.71 | 92.6991 | A1” |
| 2 | 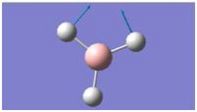
|
1204.66 | 12.3789 | E’ |
| 3 | 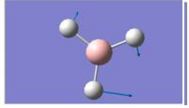
|
1204.66 | 12.3814 | E’ |
| 4 | 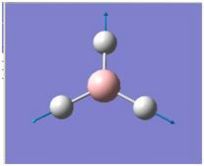
|
2592.79 | 0 | A1’ |
| 5 | 
|
2731.31 | 103.837 | E” |
| 6 | 
|
2731.31 | 103.83 | E” |
Molecular Orbitals
Gaussian was also used to observe the molecular orbitals in the BH3 molecule. Firstly a MO diagram was created for the molecule:
 The Molecular orbitals for the occupied orbitals and LUMO were calculated
The Molecular orbitals for the occupied orbitals and LUMO were calculated
HOMO (1e'):
LUMO(1a"2:
Orbital 1(:
Orbital 2:
Orbital 3:
Orbital 4:
BCl3
The same optimisation was run for BCl3 so that the differences between it and BH3 could be explored. This gave the same information as before:
| Optimised B-H bond distance | 1.86592A |
| Optimised H-B-H angle | 120° |
| File type | .log |
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Base set | LANL2MB |
| Final energy | -69.43928112au |
| Dipole moment | 0.00005905 Debye |
| Point group | D3H |
| Length of time | 13 seconds |
This molecule took a few seconds longer to calculate, probably because it is a slightly bigger molecule. The final energy and the optimised bond distance were greater in BCl3 than in BH3. This is because a greater dipole moment is created and a large molecule has been added (the chlorine) and so can not sit as closely to the Boron.
Small Molecule- H2O2
The method learnt from optimising BCl3 and BH3
was put into practice on another small molecule-H2O2.[2]
| Optimised H-O bond distance | 1.03771A |
| Optimised O-O bond distance | 1.47299A |
| Optimised H-O-O angle | 97.002° |
| File type | .log |
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Base set | LANL2MB |
| Final energy | -149.45059613au |
| Dipole moment | 0.0003 Debye |
| Point group | C2 |
| Length of time | 37 seconds |
The coordinates for this optimisation were as follows:
| Centre
Number |
Atomic
Number |
Atomic
Type |
Coordinates (Angstroms) | ||
| X | Y | Z | |||
| 1 | 8 | 0 | 0.000000 | 0.000000 | 0.000000 |
| 2 | 1 | 0 | 0.000000 | 0.000000 | 0.960000 |
| 3 | 8 | 0 | 1.244287 | 0.000000 | -0.440625 |
| 4 | 1 | 0 | 1.244287 | -0.000175 | -1.400625 |
Cis vs Trans Isomer
The cis and trans forms of the molecule Mo(CO)4(PPh3)2 were optimised using increasingly more accurate basis sets and pseudo-potentials do give a more accurate geometry optimisation.
The B3LYP Method with LANL2MB pseudo-potential was used to give a rough optimisation and then the same method but with a LANL2DZ pseudo-potential was used to give a more accurate optimisation.
Trans Structure
Trans structure |
Trans Optimised Bond Lengths and Angles [3]
| Distance/angle | Experimental | LANL2DZ Pseudo-Potential | Literature |
| Mo-C | 1.98571 A | 2.0299 A | 2.016 A |
| C=O | 1.20328 A | 1.1874 | 1.164 A |
| Mo-P | 2.81901 A | 2.61873 | 2.500 A |
| P-Ph | 1.96335 A | 1.897, 1.894, 1.892 | 1.854 A |
| P-Mo-C | 91.592° | 114.9, 117.4, 116.3 | 87.2° |
| C-Mo-C | 86.948° | 179.835 | 180° and 92.1° |
| Mo-P-Ph | 108.625°, 126.095° and 117.426 | 114.876, 116.283, 117.378 | |
| Ph-P-Ph | 102.4°, 98.257° | 103.456, 100.653 |
Cis Isomer
Transstructure |
Cis Optimised Bond Lengths and Angles
| Distance/angle | LANDL2MB Pseudo-Potential | LANL2DZ Pseudo-Potential | Literature |
| Mo-C | 2.04592 A | 1.97156 A | 2.007 A |
| C=O | 1.20367 A | 1.19118 A | |
| Mo-P | 2.68694 A | 2.73616, 2.76 A | 2.576, 2.577 A |
| P-Ph | 1.9673 A | 1.89 A | |
| P-Mo-C | 88.209° | 91.794° | 92.1° |
| C-Mo-C | 179.885° and 89.736° | 175.173° and 86.098° | 92.1° |
| Mo-P-Ph | 116.562° | 122.431 | |
| Ph-P-Ph | 100.206° | 97.664 |
The main difference between the cis and the trans isomers is the in the angles because of the different configurations.
Also obtained from the Gaussian calculations is the energy of each isomer;
| Isomer | Energy, au |
| Cis | -1923.54175254 |
| Trans | -1923.54028665 |
This shows that cis form is slightly more stable, this could be because the large PPh3 groups are further away from each other than in the cis isomer. Therefore there is less strain from these large groups. If there were a smaller group, for example a methyl group,instead of the bulky PPh3 then this would lower the strain that would occur in both isomers. If there still needed to be the bulky groups, the phenyl groups on the PPh3 could be adjusted. From the Gaussian calculation, the phenyl groups are orientated in a "windmill-like" configuration, this keeps the phenyl groups away from each other, thus reducing strain but can result in the phenyls being in conflict with other groups. The seperate phenyls could be adjusted slightly to minimise strain from both other groups in the PPh3 and the other groups in the complex.
Vibrational Frequencies
The vibrational frequencies were calculated using the frequency function in Gaussian.
The cis molecule gave some negative frequencies which the trans did not. Also, the cis molecule was expected to give 4 C=O stretches as opposed to only 3 in the trans form. This is as a result of four different vibrations in the molecule- asymmetric stretching and symmetric stretching of the two groups.
Cis Isomer [4]
| Calculation, cm-1 | Literature, cm-1 |
| 1847.77 | 1882 |
| 1851.31 | 1903 |
| 1875 | 1917 |
| 1966 | 2022 |
Trans Isomer From literature[5] it was predicted that there would be only 3 C=O stretches, probably due there being PPh3 groups in only one plane- the trans position.
| Calculation, cm-1 |
| 1842.81 |
| 1845.65 |
| 1890.65 |
| 1959(very low intensity) |
There are 4 peaks, but the last peak is at a very low intensity, this corresponded to the symmetric stretch
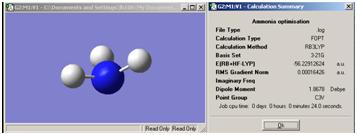
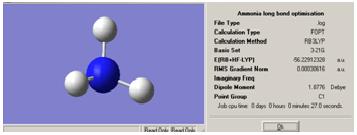
Inversion of Ammonia
First, a model of ammonia was created and optimised.
Another model was made with one N-H bond being 1.01A long rather than 1A. This changes the symmetry of the molecule from C3V to C1
Another model with D3H symmetry was optimised using the same method:

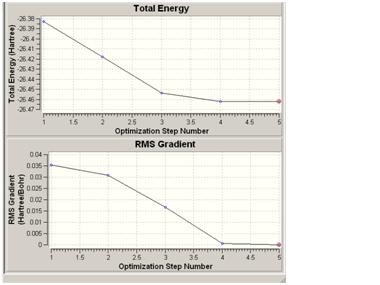
The structure of the D3H molecule differed from the other two in that the H-N-H angle was 120° as opposed to 110° in the other symmetries. This is to fit in the D3H symmetry model. The symmetry of the molecule also had an effect on the time taken to optimise the geometry. The more the molecule was altered from the original C3V model, the longer it took for the calculation to run. This suggests that the symmetry closest to the real situation is the quickest to optimise and so will give the quickest calculation time. Normally the molecule cannot break symmetry during an optimisation but, especially in the case of high symmetry molecules, the symmetry can be "turned off". This inability to break symmetry has ramifications for highly symmetrical molecules as they may need to break symmetry. If they cannot, the most optimised form may not be found.
The lowest energy geometry was for the D3H symmetry optimisation with -56.53154189 au. Although all three geometries gave very similar energies as they are effectively the same molecule. The energies for the other isomers are as follows: C3V= -2.14165496E-2KJ/mol-1, C1=-2.14165485E-2KJ/mol-1. Hence the difference between these energies and the lowest D3H energy can be calculated as:C3V= 1.1518407E-4, C1=1.1518520E-4. This difference does seem small but could still be significant.
The C3V and D3H models were optimised further using the 6-311+G(d,p) basis set so that the inversion of ammonia could be observed.
The calculations using this basis set took longer than for its lower level counterpart. This is probably because this is a more accurate calculation and hence requires more time to complete.
The change in energy between the C3V and D3H geometries was calculated (the process of the inversion). This was found to be 2.96934E-6. The ΔE value has therefore been reduced between the two basis sets by the use of a more accurate basis set. These results are a lot lower than the expeirmentally determined barrier of 24.3 KJ/mol, this could be because the computational method gives a more accurate value.
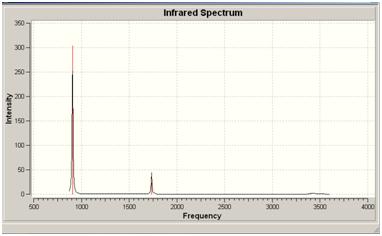
Ammonia Vibrational Frequencies
The frequencies of the C3V and D3H models were calculated using gaussian and compared.
| C3V | Calculated | Literature | Matched up? | D3H | Calculated | |
| Frequency | Intensity | Frequency | Intensity | Frequency | Intensity | |
| 906.325 | 303.409 | 1139 | 1 | Yes | -318.051 | 849.113 |
| 1734.71 | 22.6457 | 1765 | 1.1E-6 | Yes | 1640.59 | 55.9805 |
| 1734.71 | 22.6456 | 1765 | 1.1E-6 | Yes | 1640.59 | 55.9828 |
| 3401.89 | 2.1217 | 3464 | 0.073 | Yes | 3635.71 | 0 |
| 3566.88 | 0.0635 | 3534 | 0.052 | Yes | 3854.27 | 19.5963 |
| 3566.88 | 0.0634 | 3646 | 0.073 | Yes | 3854.71 | 19.5951 |
The C3H geometry gave 6 positive frequencies with two pairs of degenerate levels. The D3H isomer gave only 5 positive frequencies, again with two degenerate pairs. As the D3H structure is a transition state it has a negative frequency, this occured at -318.051 whereas the C3V,being a ground state structure does not have this and only has positive frequencies.
The first vibrational mode (pictured below) follows the reaction path as it stays the same for both symmetries.

References
- ↑ Advanced Organic Chemistry, Francis A. Carey, Richard J. Sundberg
- ↑ DOI:10042/to-1358
- ↑ DOI:10042/to-1231
- ↑ DOI:10042/to-1238
- ↑ http://article.pubs.nrc-cnrc.gc.ca/RPAS/rpv?hm=HInit&calyLang=eng&journal=cjc&volume=46&afpf=v68-276.pdf A. D. Allen. and P. F. Barrett, Candian Journal of Chemistry, 1968, 46, 1649