Rep:Mod:lc2.25.19
Molecular Modelling 2
Molecules examined in this workshop: NO3, H2, N2, PF5,
NH3
Calculation method: B3LYP
basis set: 6-31G(d.p)
final energy: -56.55776873 a.u.
RMS gradient: 0.00000485 a.u.
point group: C3v
Bond length: 1.018Å
Bond angle: 105.7°
| Item | Value | Threshold | Converge? | |
|---|---|---|---|---|
| Maximum | Force | 0.000004 | 0.000450 | YES |
| RMS | Force | 0.000004 | 0.000300 | YES |
| Maximum | Displacement | 0.000072 | 0.001800 | YES |
| RMS | Displacement | 0.000035 | 0.001200 | YES |
Log File: File:CD1618NH3result.LOG
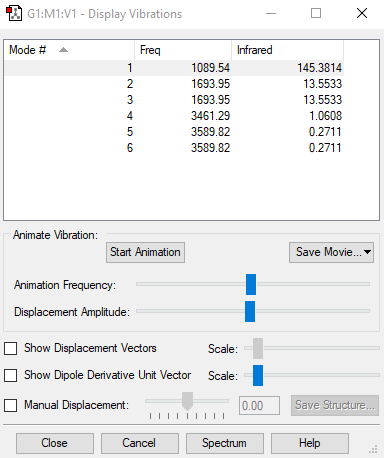
| Mode | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| symmetry | A1 | E | E | A1 | E | E |
| Frequencies | 1090 | 1694 | 1694 | 3461 | 3590 | 3590 |
| IR Inten | 145 | 14 | 14 | 1 | 0 | 0 |
NH3 |
Charge distribution: N: -1.125 H: 0.375
Questions
1. 3*4-6=6 so 6 modes
2. two pairs of vibration modes with frequency 1694 and 3590
3. Bond stretch: 3461cm-1 3590cm-1 3590cm-1 Bending 1090cm-1 1694cm-1 1694cm-1
4. 1090cm-1 3461cm-1
5. 1090cm-1
6. 2 bands since there are six vibration modes
H2
Calculation method: B3LYP
basis set: 6-31G(d.p)
final energy: -1.17853935 a.u.
RMS gradient: 0.00003809 a.u.
point group: D∞h
Bond length: 0.743Å
| Item | Value | Threshold | Converge? | |
|---|---|---|---|---|
| Maximum | Force | 0.000066 | 0.000450 | YES |
| RMS | Force | 0.000066 | 0.000300 | YES |
| Maximum | Displacement | 0.000087 | 0.001800 | YES |
| RMS | Displacement | 0.000123 | 0.001200 | YES |
Log File: File:CD1618H2OUTPUT.LOG
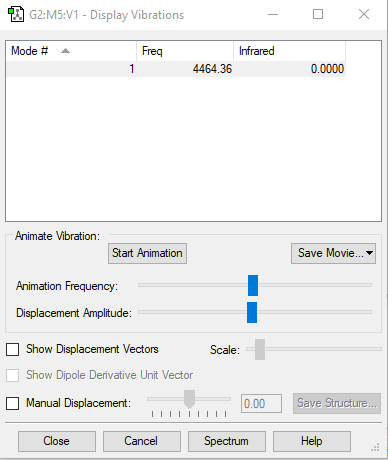
| Mode | 1 |
|---|---|
| symmetry | SGG |
| Frequencies | 4464 |
| IR Inten | 0 |
H2 |
Identifier for the complex: CILXAX
Bond length in the complex: 0.8Å
N2
Calculation method: B3LYP
basis set: 6-31G(d.p)
final energy: -109.52412868 a.u.
RMS gradient: 0.00000365 a.u.
point group: D∞h
Bond length: 1.106Å
| Item | Value | Threshold | Converge? | |
|---|---|---|---|---|
| Maximum | Force | 0.000006 | 0.000450 | YES |
| RMS | Force | 0.000006 | 0.000300 | YES |
| Maximum | Displacement | 0.000002 | 0.001800 | YES |
| RMS | Displacement | 0.000003 | 0.001200 | YES |
Log File: File:CD1618N2OUTPUT.LOG
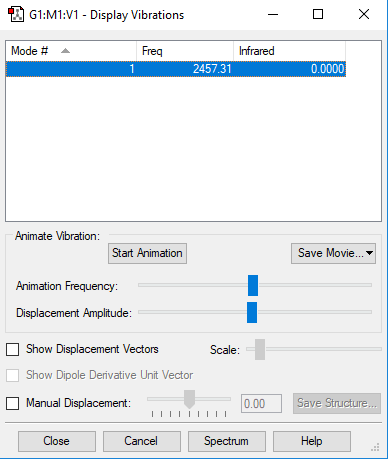
| Mode | 1 |
|---|---|
| symmetry | SGG |
| Frequencies | 2457 |
| IR Inten | 0 |
N2 |
Identifier for the complex: VEJFOI
Bond length in the complex: 1.092Å
In the complex, the bond length of N2 and H2 is longer than the calculated value. Theoretically, when forming a dative bond, the electron from the metal is feed into the LUMO of the ligand molecule, so the bond order of ligand molecule decreases and the anti-bonding character increases. Thus a longer and weaker bond is observed.
Energy in Harbor Process
E(NH3)= -56.55776873 a.u.
2*E(NH3)= -113.1155375 a.u.
E(N2)= -109.52412868 a.u.
E(H2)= -1.17853935 a.u.
3*E(H2)= -3.53561805 a.u.
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -146.4785616 kJ/Mol
The ammonia product is more stable.
PF5
Calculation method: B3LYP
basis set: 6-31G(d.p)
final energy: -109.52412868 a.u.
RMS gradient: 0.00000365 a.u.
point group: D3h
axial P-F bond length: 1.597Å
planar P-F bond length: 1.569Å
F-P-F bond angle: 90°
F-P-F bond angle: 120°
| Item | Value | Threshold | Converge? | |
|---|---|---|---|---|
| Maximum | Force | 0.000299 | 0.000450 | YES |
| RMS | Force | 0.000090 | 0.000300 | YES |
| Maximum | Displacement | 0.000868 | 0.001800 | YES |
| RMS | Displacement | 0.000269 | 0.001200 | YES |
Log File: File:CD1618PF5INPUT.LOG
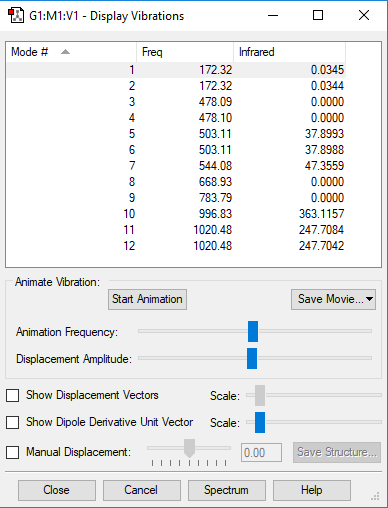
| Mode | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| symmetry | E' | E" | E" | E" | E' | E' | A2" | A1' | A1' | A2" | E' | E' |
| Frequencies | 172 | 172 | 478 | 478 | 503 | 503 | 544 | 669 | 784 | 997 | 1020 | 1020 |
| IR Inten | 0 | 0 | 0 | 0 | 38 | 38 | 47 | 0 | 0 | 363 | 248 | 248 |
PF5 |
Charge on F: -0.578
Charge on P: 2.748
MO
Axial F-P-F is z axis in the disciession below
-0.47310 a.u.
Three 2p orbitals from equatorial F atoms, there is three nodal planes and each orbital is out of phase with the orbital next to it.
-0.45487 a.u.
Three 2pz orbitals from equatorial F atoms art in phase and two axial 2pz orbitals are out of phase with each other and equatorial orbitals.
-0.43685 a.u.
Three 2p orbitals from equatorial F atoms are interacting, but the electron density is mainly on the axial F atoms,and these 2p orbitals are in phase with each other.
-0.41452 a.u.
This MO has a nodal plane going across the P-F bond and all the 2p orbitals from the other F atoms are out of phase.
-0.41452 a.u.
A pair of 2pz orbitals from equatorial F atoms art in phase but out of phase with the last one and two axial 2pz orbitals are out of phase with each other and equatorial orbitals. This is the HOMO
0.00039 a.u.
The 4s orbial of central P is out of phase with each of the 2p orbital from F atoms. THis is the LUMO
Marking
Note: All grades and comments are provisional and subject to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have received your grade from blackboard.
Wiki structure and presentation 1/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES
NH3 0.5/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
You didn't answer the question asking you to explain what you would expect the charge distribution to be in ammonia. Your answer to question 6 doesn't explain why there are only 2 modes - there are two sets of degenerate vibrations which would give 4 peaks, but 2 of these are too low in intensity to be visible experimentally.
N2 and H2 0.5/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES - good explanation, well done.
Haber-Bosch reaction energy calculation 0.5/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
No, you should have reported to 1 d.p. maximum.
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 3/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
YES - however you could have explained the charges in terms of the relative electronegativity of the elements. Your MO descriptions are good but lacking in detail.
Independence 0/1
If you have finished everything else and have spare time in the lab you could: Check one of your results against the literature, or Do an extra calculation on another small molecule, or Do some deeper analysis on your results so far