Rep:Mod:kutyalo 96
NH3 Molecule
Key Information
Molecule - NH3
Calculation Method - RB3LYP
Basis Set - 6-31G(d,p)
Final Energy E(RB3LYP) - -56.55776873
RMS Gradient - 0.0000485
Point Group - C3V
Optimised Bond Distance - 1.01798 au
H-N-H Bond Angle - 105.741 degree
Item Table
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES Predicted change in Energy=-5.986278D-10 Optimization completed.
Jmol Dynamic Image
test molecule |
Vibrations
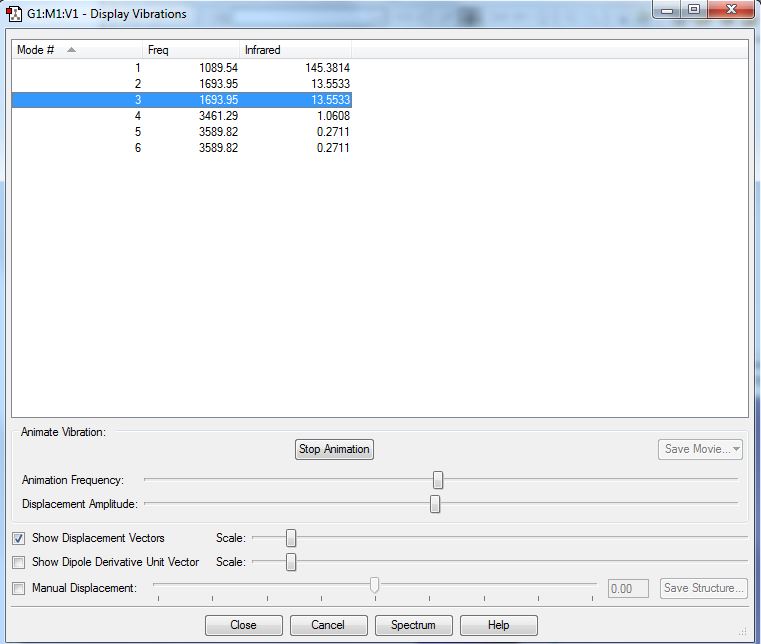
According to the 3N-6 rule, I would expect to have 12-6=6 modes.
Modes 2 and 3; 5 and 6 are degenerate.
Bending modes: 1,2,3; Stretching modes - 4,5,6
Mode number 4 is highly symmetric.
Mode number 1 is known as the umbrella mode.
I would expect to see 3 bands in the experimental spectrum of gaseous ammonia, as it has 3 different energy bands.
Charges in Ammonia
Charge on N = -1.125
Charge on H = 0.375
I would expect to have a negative charge on the N atom, as it has a higher value of electronegativity.
N2 Molecule
Key Information
Calculation Method - RB3LYP
Basis Set - 6-31G(d,p)
Final Energy E(RB3LYP) - -109.52412868
RMS Gradient - 00000060
Point Group - D*H
Item Table
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.401024D-13
Optimization completed.
-- Stationary point found.
Jmol Dynamic Image
test molecule |
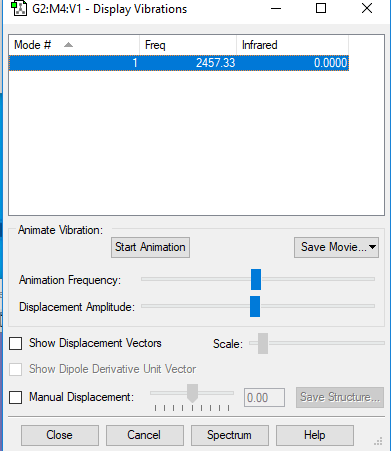
Vibrations
1 mode, frequency - 2457.33; IR - 0.00
H2 Molecule
Key Information
Calculation Method - RB3LYP
Basis Set - 6-31G(d,p)
Final Energy E(RB3LYP) - -1.17853936
RMS Gradient - 0.00000017
Point Group - D*H
Bond Lenght - 0.74279 au
Item Table
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES Predicted change in Energy=-1.164080D-13 Optimization completed.
Jmol Dynamic Image
test molecule |
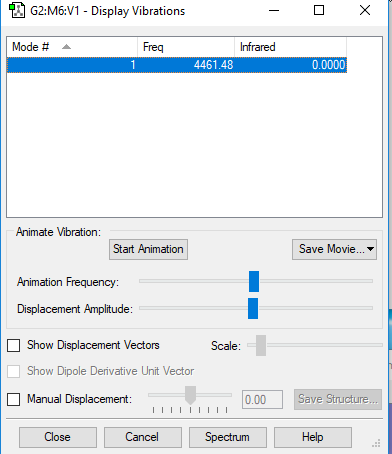
Vibrations
Only 1 mode.
NH3 Synthesis
E(NH3)= -56.55776873 au
2*E(NH3)= -113.115537 au
E(N2)= -109.52412868 au
E(H2)= -1.17853936 au
3*E(H2)= -3.53561808 au
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -146.47727512 KJ/mol
Reactants are less stable
CH4 Molecule
Key Information
Calculation Method - RB3LYP
Basis Set - 6-31G(d,p)
Final Energy E(RB3LYP) - -40.52401404
Gradient - 0.00003263
Point Group - TD
Item Table
Item Value Threshold Converged?
Maximum Force 0.000063 0.000450 YES
RMS Force 0.000034 0.000300 YES
Maximum Displacement 0.000179 0.001800 YES
RMS Displacement 0.000095 0.001200 YES
Predicted change in Energy=-2.256034D-08
Optimization completed.
-- Stationary point found.
Jmol Dynamic Image
test molecule |
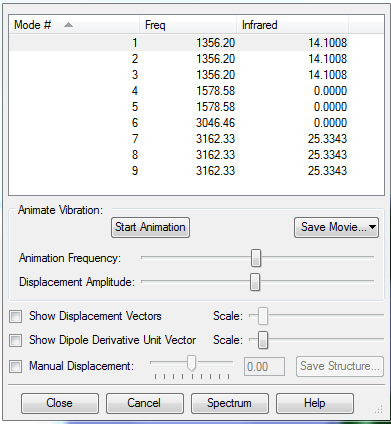
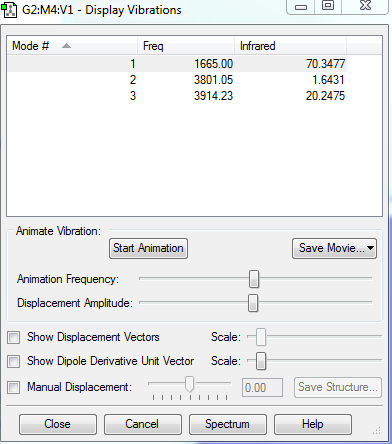
Vibrations
According to the 3N-6 rule, I would expect to have 15-6=9 modes.
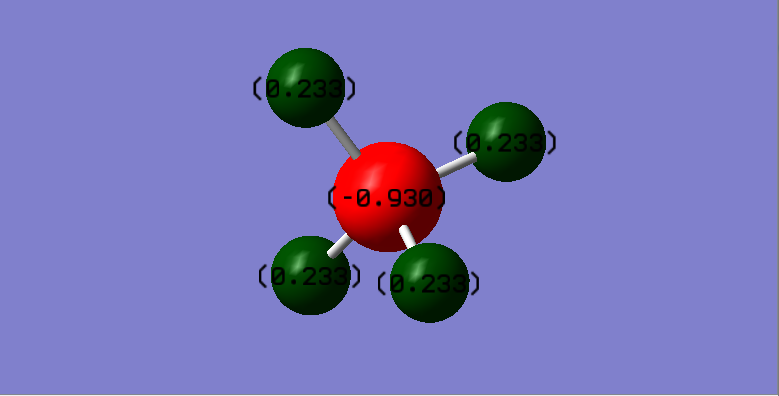
Charge Distribution
Atomic charges:
C - -0.930
H - 0.233
Molecular Orbitals
–Derived from the 1s atomic orbital of the C atom.
–Core orbital, thus deep in energy.
–Non-bonding, occupied by 2 electrons.
–Have no effect on bonding.
–Hybridised molecular orbital, combination of 2s+2p electrons from C and 1s electrons from H.
–Deeper in energy than other hybridised molecular orbitals due to increased s character.
–Differs from an s molecular orbital because its slightly directional.
–Bonding, sigma bond between C and H, occupied by 2 electrons.
–3 identical hybridised molecular orbital (pointing in different directions)
–HOMO, energy in the HOMO/LUMO region.
–Mixture of 2s+2p electrons from C and 1s electrons from H
–Bonding, sigma bond between C and H, occupied by 2 electrons.
–Higher in E than the first hybridised molecular orbital.
–There are 3 degenerate bonding orbitals at this E level.
–Corresponding to the first bonding molecular orbital (from 1s on C)
–LUMO, its energy is therefore in the HOMO/LUMO region.
–Unoccupied antibonding orbital.
–Unfilled, has no contribution to bonding.
–Lower in E than other hybridised antibonding orbitals, thus harder to occupy (harder to break this bond)
–Corresponding to hybridised molecular orbital which was lower in E due to different s contribution.
–Unoccupied, antibonding orbital.
–Its lower in Energy than other hybridised antiobonding orbitals thus harder to occupy.
–Unfilled, has no contribution to bonding.
–There are 3 degenerate antibonding orbitals at this E level.
Independence Section - H2O Molecule
Key Information
Calculation Method - RB3LYP
Basis Set - 6-31G(d,p)
E(RB3LYP) - -76.4197374 au
RMS Gradient Norm - 0.00006276 au
Dipole Moment - 2.0428 debye
Point Group - C2V
Bond Lenght - 0.96522 au
Bond Angle - 103.745 degree
Item Table
Item Value Threshold Converged?
Maximum Force 0.000099 0.000450 YES
RMS Force 0.000081 0.000300 YES
Maximum Displacement 0.000115 0.001800 YES
RMS Displacement 0.000120 0.001200 YES
Predicted change in Energy=-1.939669D-08
Optimization completed.
-- Stationary point found.
Jmol Dynamic Image
test molecule |
Vibrations
According to the 3N-6 rule, I would expect to have 9-6=3 modes.