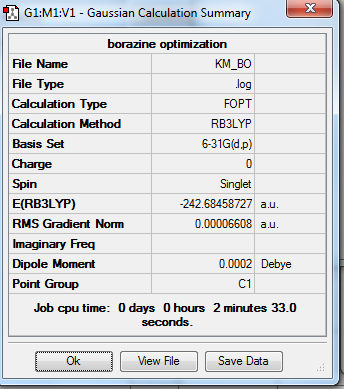
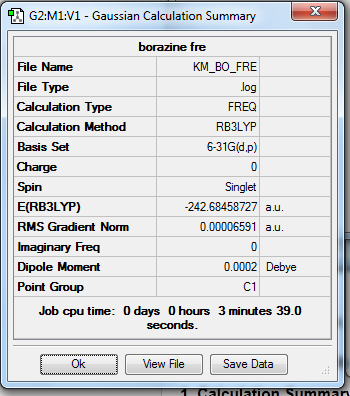
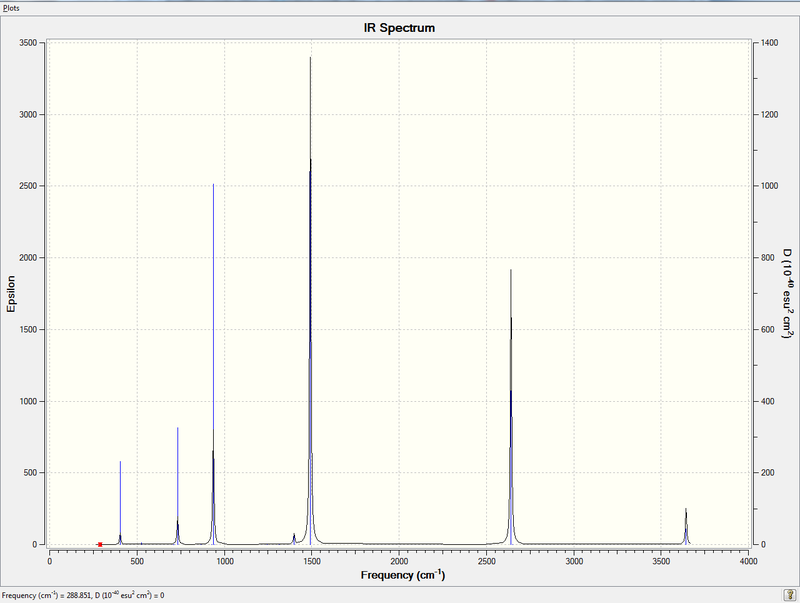
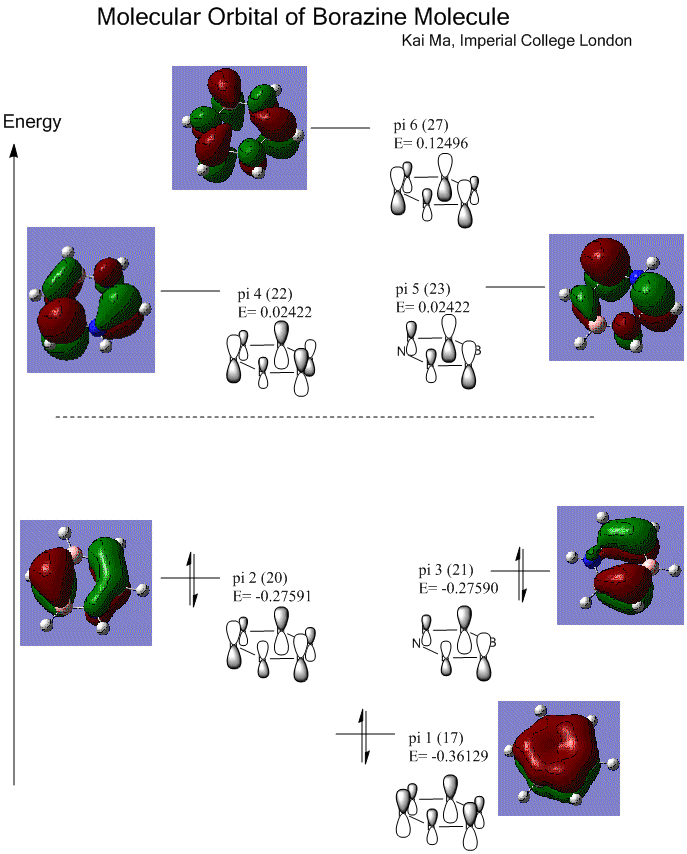
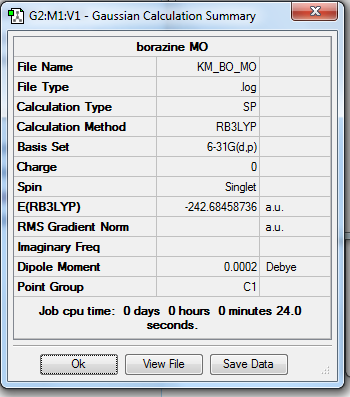
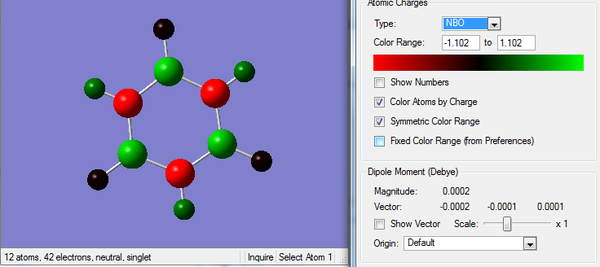
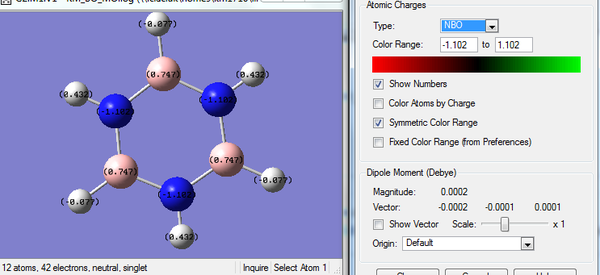
Rep:Mod:km1710
Optimization Analysis
Optimization of BH3 (using 3-21G basis set)
1. Optimized BH3 Molecule Using 3-21G Basis Set
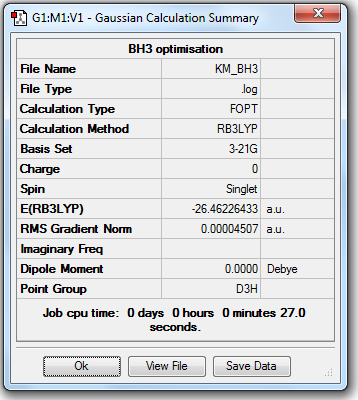
Initially, the molecule BH3 was set to a trigonal planar structure and bond angle of 120.0o between each B-H bond with bond length 1.5 Å. What the program did was to make assumption of the nuclei at different position and with varied geometries, compare between each situation and decide the structure with lowest energy. In the first stage, B3LYP method and 3-21G basis set was used for optimization.
test molecule |
2. Geometry Data
Bond length for each B-H: 1.1945 Å
Bond angel between each B-H: 120.0o
3. Real Output of Optimised BH3 molecule
Item Value Threshold Converged?
Maximum Force 0.000090 0.000450 YES
RMS Force 0.000059 0.000300 YES
Maximum Displacement 0.000352 0.001800 YES
RMS Displacement 0.000230 0.001200 YES
Predicted change in Energy=-4.580970D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1945 -DE/DX = -0.0001 !
! R2 R(1,3) 1.1945 -DE/DX = -0.0001 !
! R3 R(1,4) 1.1945 -DE/DX = -0.0001 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
4. Calculation Summary
5. File Link
To access to the file of optimization, click here.
Optimization of BH3 (using 6-31G(d,p) basis set)
1. Optimized BH3 Molecule Using 6-31G(d,p) Basis Set
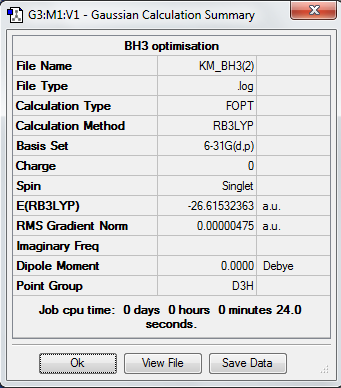
Another optimization based on a more accurate basis set, 6-31G(d,p) was carried out and results are shown as following:
test molecule |
2. Geometry Data
Bond length for each B-H: 1.1923 Å
Bond angel between each B-H: 120.0o
3. Real Output of Optimised BH3 molecule
Item Value Threshold Converged?
Maximum Force 0.000006 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000023 0.001800 YES
RMS Displacement 0.000015 0.001200 YES
Predicted change in Energy=-2.008855D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1923 -DE/DX = 0.0 !
! R2 R(1,3) 1.1923 -DE/DX = 0.0 !
! R3 R(1,4) 1.1923 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
4. Calculation Summary
5. File Link
To access to the file of optimization, click here.
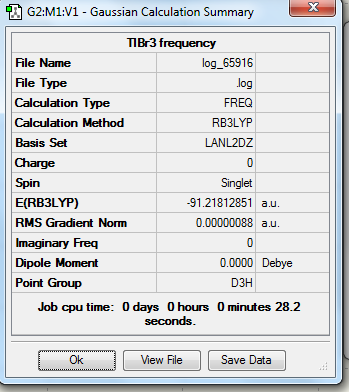
Optimization of TlBr3 (using pseudo potentials and LanL2DZ basis set)
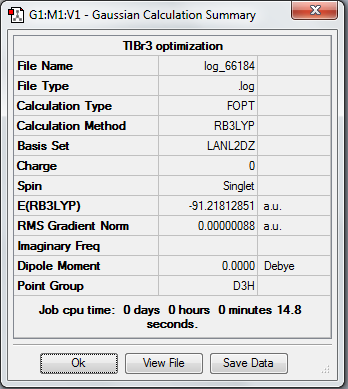
1. Optimized TlBr3 molecule using pseudo potentials and LanL2DZ basis set
In this molecule, LanL2DZ basis set was carried out as following.
test molecule |
2. Geometry Data
Bond length for each B-H: 2.651 Å
Bond angel between each B-H: 120.0o
3. Real Output of Optimised TlBr3 molecule
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000016 0.001800 YES
RMS Displacement 0.000010 0.001200 YES
Predicted change in Energy=-4.107304D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.651 -DE/DX = 0.0 !
! R2 R(1,3) 2.651 -DE/DX = 0.0 !
! R3 R(1,4) 2.651 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
4. Calculation Summary
5. File Link
To access to the D-space link, click | here.
Optimization of BBr3 (using pseudo potentials and mixture of basis sets)
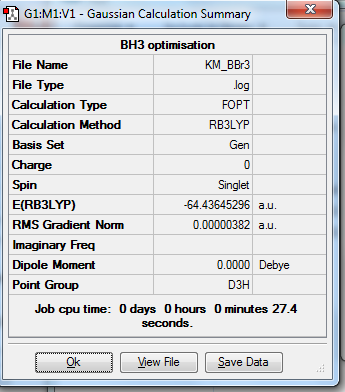
1. Optimized BBr3 molecule using pseudo potentials and mixture of basis sets
In this molecule, a mixture of basis sets were used in order to obtain a more accurate optimization. This is due to the involvement of the mixture of heavy atoms (Br) and light atom (B). 6-31G(d,p) basis set that directed from the optimized BH3 molecule was used for B atom and LanL2DZ basis set was used for Br atoms.
test molecule |
2. Geometry Data
Bond length for each B-H: 1.9340 Å
Bond angel between each B-H: 120.0o
3. Real Output of Optimised BBr3 molecule
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000036 0.001800 YES
RMS Displacement 0.000023 0.001200 YES
Predicted change in Energy=-4.027684D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.934 -DE/DX = 0.0 !
! R2 R(1,3) 1.934 -DE/DX = 0.0 !
! R3 R(1,4) 1.934 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
4. Calculation Summary
5. File Link
To access to the D-space Link, click here.
Result and Discussion
1. Comparison of Bond Lengths
| Molecule | Bond length/ Å | Literature Bond Length/ Å |
| BH3 (3-21G) | 1.1945 | 1.19[1] |
| BH3 (6-31G) | 1.1923 | 1.19 |
| BBr3 | 1.9334 | Not Found |
| TlBr3 | 2.651 | 2.564[2] |
Bond length refers to the average distance between two centres of atoms connected via a chemical bond, which origins from the attraction between two atoms. It is the result from electron sharing in a covalent bond. Factors affecting bond length include size of atom, electronegativity of atom and efficiency of electron overlap. In this experiment, the calculated bond lengths of BH3 matches well with literature value. It also demonstrates that the 6-31G(d,p) basis set is more accurate than the 3-21G basis set because the resulted value is more close to literature. The optimized TlBr3 bond length, however, shows a slightly higher value than literature because a medium level basis set was used in calculation. In order to get more accurate results, higher level of basis set must be proceeded in this molecule.
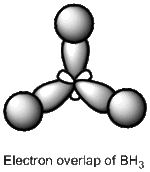
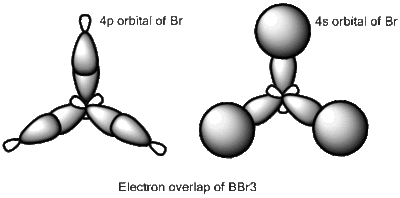
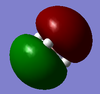
BBr3 molecule shows a longer B-Br bond than the B-H bond in BH3 molecule. There are two reasons responsible for the difference: electronegativity and atom size. Br is more electronegative atom than H atom, therefore pulls more electron density from B atom. The greater size of Br atom also results in less effective overlap of electron overlap, hence larger bond length. A brief image of how the electrons have been overlapped in these two molecules is shown below.
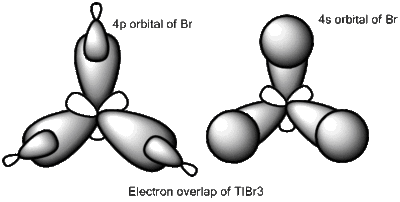
When comparing bond lengths between B-Br bond in BBr3 and Tl-Br bond in TlBr3, the difference becomes even larger due to the large difference between atomic sizes of Tl and B. Both Tl and B are in group 13, thus have very similar molecular structures in TlBr3 and BBr3. The bond tends to be much weaker and longer between Br-Tl and the overlap becomes even poorer. The diagram of electron overlap of TlBr3 is shown as following.
Both B and Tl in these molecules are sp2 hybridized.
In Guassview, sometimes the bond is not drawn because the program works on a distance criteria which is usually pre- defined. Chemical bonds are drawn when there is considered to be high electron density in a specific position. Therefore, when the calculated electron density is too low or distance exceeds the pre- defined value, no bond will be drawn in the program. This does mean the non- existence of bond, but in contrast, gives indication on the relatively weakness of bond and less effective overlap.
2. Energy Difference in Different Basis Sets
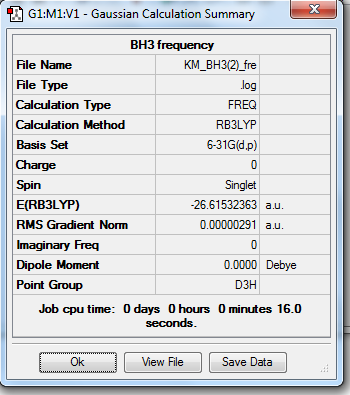
A difference in energy has been shown between optimization of BH3 molecule using different basis sets. In the 3G-21 basis set, the total energy is -26.46226433 a.u., in the 6G-31(d,p) basis set, the total energy is -26.61532363 a.u.
The difference in total energy is therefore: 0.1530593 a.u., i.e., 401.8572 kJ/mol. The energy difference is quite high and indicates that it makes no sense comparing between different basis sets.
3. Symmetry and Dipole Moments
All the optimized molecules have the correct symmetry (D3h) and zero net dipole moments. This corresponds to literature well and proves that the optimization is relatively accurate.
Frequency Analysis
Frequency Analysis of BH3
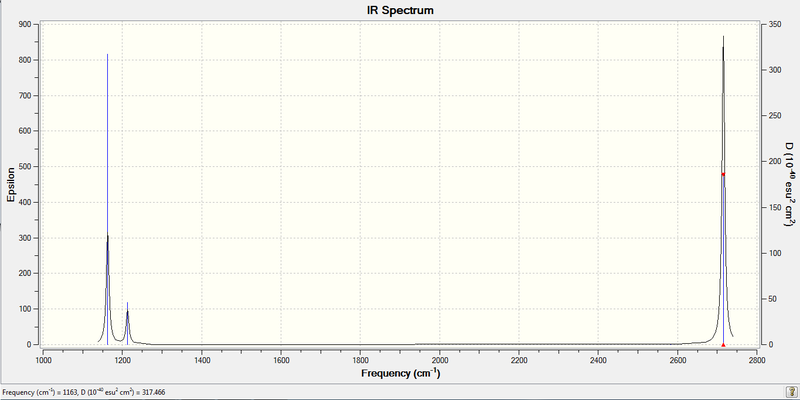
1. Real Output of Frequency Analysis of BH3
Low frequencies --- -0.9432 -0.8611 -0.0054 5.7455 11.7246 11.7625
Low frequencies --- 1162.9963 1213.1826 1213.1853
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A2" E' E'
Frequencies -- 1162.9963 1213.1826 1213.1853
Red. masses -- 1.2531 1.1072 1.1072
Frc consts -- 0.9986 0.9601 0.9601
IR Inten -- 92.5482 14.0551 14.0587
Atom AN X Y Z X Y Z X Y Z
1 5 0.00 0.00 0.16 0.00 0.10 0.00 -0.10 0.00 0.00
2 1 0.00 0.00 -0.57 0.00 0.08 0.00 0.81 0.00 0.00
3 1 0.00 0.00 -0.57 0.39 -0.59 0.00 0.14 -0.39 0.00
4 1 0.00 0.00 -0.57 -0.39 -0.59 0.00 0.14 0.39 0.00
2. Calculation Summary
After calculation, the total energy, dipole moment and symmetry group stays exactly the same as before.
3. Vibration Modes for BH3
| No. | Vibration Mode | Description | Frequency/ cm-1 | Literature Frequency/ cm-1[3] | Intensity | Symmetry to D3h Point Group |
|---|---|---|---|---|---|---|
| 1 |  |
H atoms moves in a plane concertedly while B atom moves in an opposite direction. This movement results in an increase of dipole moment to a great extent and therefore IR active. i.e., large intensity. | 1163.00 | 1163 | 92.5482 | A2' |
| 2 | 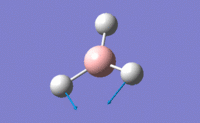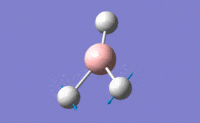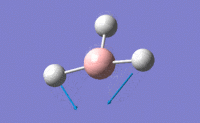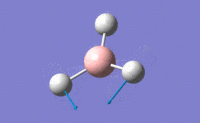 |
Only two H atoms scissor while the other H atom and central B atom remain unchanged in position. This results in only a small change in dipole moment, thus small value of intensity. | 1213.18 | 1223 | 14.0551 | E' |
| 3 | 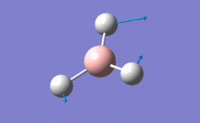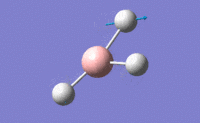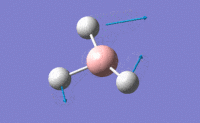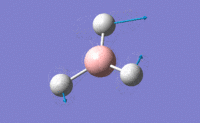 |
Two H atoms move along the plane and the other H atom is wagging in opposite direction. B atom moves slightly with the movement of H atoms. The resultant intensity is relatively small. | 1213.19 | 1223 | 14.0587 | E' |
| 4 |  |
All three H atoms vibrate in and out in the same plane concertedly and B atom stays unchanged in position. The net dipole moment is zero, thus no intensity. | 2582.28 | 2567 | 0.0000 | A1' (totally symmetric) |
| 5 |  |
Two H atoms stretch in same plane but in opposite directions while the remaining H atom does not change the position. B atom also moves slightly with H atoms. This results in a large change in dipole moment. | 2715.45 | 2696 | 126.3302 | E' |
| 6 |  |
Two H atoms move in an opposite direction of the remaining H atom in a plane. B atom moves slightly with the two H atoms. The resultant intensity is large. | 2715.45 | 2696 | 126.3206 | E' |
The predicted frequency matches well with literature.
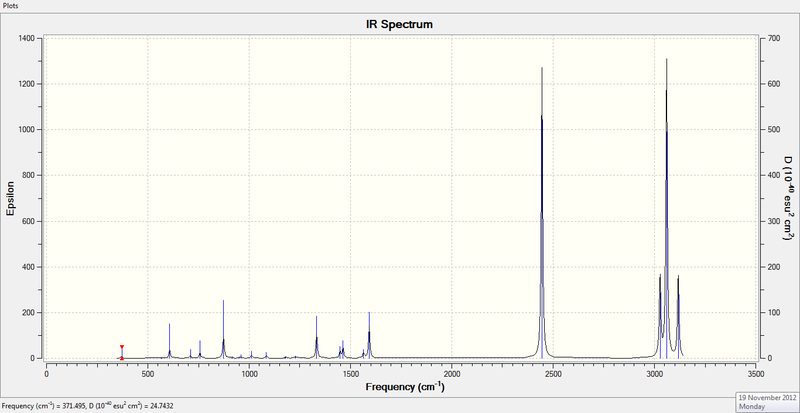
3. IR Spectrum of Optimized BH3
4. File Link
To access to the file link, click here
Frequency Analysis of TlBr3
Frequency analysis of optimized TlBr3 molecule was investigated without any change in basis set.
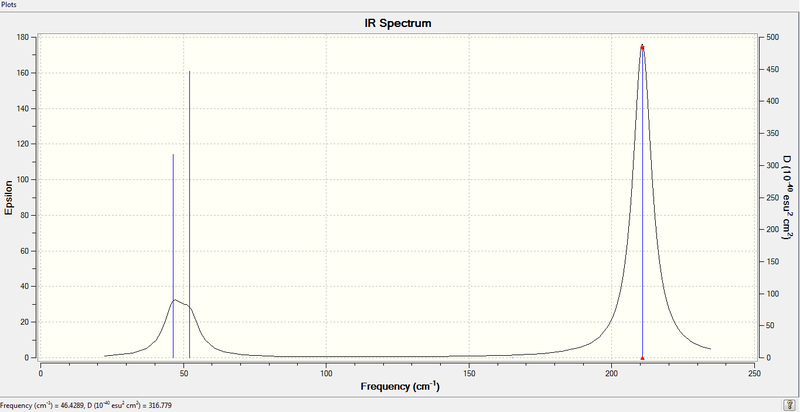
1. Real Output of Frequency Analysis of TlBr3
Low frequencies --- -3.4213 -0.0026 -0.0004 0.0015 3.9367 3.9367
Low frequencies --- 46.4289 46.4292 52.1449
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
E' E' A2"
Frequencies -- 46.4289 46.4292 52.1449
Red. masses -- 88.4613 88.4613 117.7209
Frc consts -- 0.1124 0.1124 0.1886
IR Inten -- 3.6867 3.6867 5.8466
Atom AN X Y Z X Y Z X Y Z
1 81 0.00 0.28 0.00 -0.28 0.00 0.00 0.00 0.00 0.55
2 35 0.00 0.26 0.00 0.74 0.00 0.00 0.00 0.00 -0.48
3 35 0.43 -0.49 0.00 -0.01 -0.43 0.00 0.00 0.00 -0.48
4 35 -0.43 -0.49 0.00 -0.01 0.43 0.00 0.00 0.00 -0.48
2. Calculation Summary
After calculation, the total energy, dipole moment and symmetry group stays exactly the same as before.
3. Vibration Modes for TlBr3
| No. | Vibration Mode | Description | Frequency/ cm-1 | Literature Frequency/ cm-1[4] | Intensity | Symmetry to D3h Point Group |
|---|---|---|---|---|---|---|
| 1 |  |
Two Br atoms scissor toward each other while Tl atom and the other Br atom move toward the opposite direction in the same plane. The resultant change in dipole moment is rater small. | 46.43 | 47 | 3.6867 | E' |
| 2 |  |
Very similar movement to vibration mode No.1 but the third Br atom rocks in the plane rather than stretching with Tl atom. The resultant dipole moment is also small. | 46.43 | 47 | 3.6867 | E' |
| 3 |  |
All three Br atoms move concertedly in a plane while Tl atom is wagging in an opposite direction. This results in a small change in dipole moment as well. | 52.14 | 63 | 5.8466 | A2' |
| 4 |  |
All three Br atoms vibrate in and out in the same plane concertedly and Tl atom stays unchanged in position. The net dipole moment is zero, thus no intensity. | 165.27 | N/A | 0.0000 | A1' |
| 5 |  |
Two Br atoms stretch in same plane but in opposite directions while the remaining Br atom does not change the position. Tl atom also moves slightly with H atoms. This results in a large change in dipole moment. | 210.69 | 203 | 25.4830 | E' |
| 6 |  |
Two Br atoms move in an opposite direction of the remaining Br atom in a plane. Tl atom moves slightly with the two Br atoms. The resultant intensity is large. | 210.69 | 203 | 25.4797 | E' |
4. IR Spectrum of TlBr3
5. File Link
To access to the D-space link, click |here.
Result and Discussion
1. Peak Counting in IR Spectra
Theoretically there is supposed to be 3N-6 peaks, which is 6 peaks in both IR spectra of BH3 and TlBr3 molecule. However, only 3 peaks were observed and some explanations can be made to rationalize this phenomenon.
In IR spectrum of BH3, firstly, in vibration mode No.4, there is no resultant change in dipole moment, therefore it is not IR active and gives zero intensity in the spectrum.
Also, vibration modes 2&3 and 5&6 both have E1' symmetry and very similar frequencies. This causes degeneration. In other words, each pair of vibration modes contributes only one peak in the spectrum.
Again, there are only 3 peaks in spectrum of TlBr3. The reason is the same as for BH3 that A1' symmetry has no intensity and each of the E' pair contributes to one peak.
2. Comparison Between Frequencies
| No. of Vibration Mode | Frequency of BH3/ cm-1 | Frequency of TlBr3/ cm-1 | Symmetry of BH3 | Symmetry of TlBr3 |
|---|---|---|---|---|
| 1 | 1163.00 | 46.43 | A2' | E' |
| 2 | 1213.18 | 46.43 | E' | E' |
| 3 | 1213.19 | 52.14 | E' | A2' |
| 4 | 2582.28 | 165.27 | A1' | A1' |
| 5 | 2715.45 | 210.69 | E' | E' |
| 6 | 2715.45 | 210.69 | E' | E' |
The frequencies of BH3 molecule are apparently much higher than frequencies of TlBr3 molecule. The first reason is that Tl-Br bond is weakerthan B-H bond as discussed previously , therefore the resultant frequency of vibrations tend to be weaker. Additionally, the reduces mass of TlBr3 (55.371) is much larger than that of BH3 (0.915). Since the vibrational frequency is proportional to reduce mass1/2, the frequency of TlBr3 tends to be small as well.
The similarity between two molecules is that both have 6 vibrational modes and 3 peaks observed in IR spectrum.
A reordering of vibrational modes is observed as in BH3, the lowest frequency has A2' symmetry but in TlBr3 molecule, the degenerate E' symmetry modes occupy the lowest frequency.
3. Theories and Background Information
3.1 Why must you use the same method and basis set for both the optimization and frequency analysis calculations?
Different basis sets lead to large difference in total energy and accuracy of results as calculation in optimization of BH3 in the very first part. It does not make any sense if comparing molecules that have been optimized or frequency analyzed using different basis sets.
3.2 What is the purpose of carrying out a frequency analysis?
Frequency analysis is the second derivative of the potential energy surface, in which positive frequencies indicate a minimum in energy and negative frequencies indicate the existence of a transition state (i.e. failure in optimization). It can also be made to compare with experimental data.
3.3 What do the "Low frequencies" represent?
If a negative 'low frequency' is obtained, it means failure in optimization that the molecule is still not in its minimum energy state. It could turn to positive value if optimization is carried out for several more times of using a more accurate basis set.
Molecular Orbital Analysis of BH3
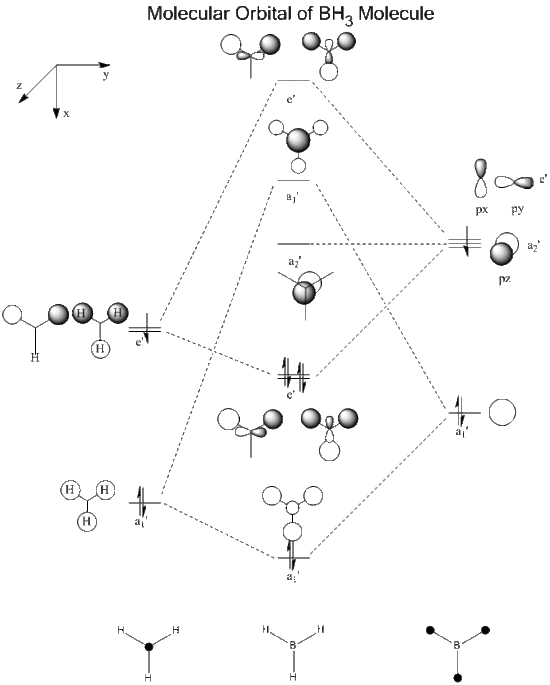
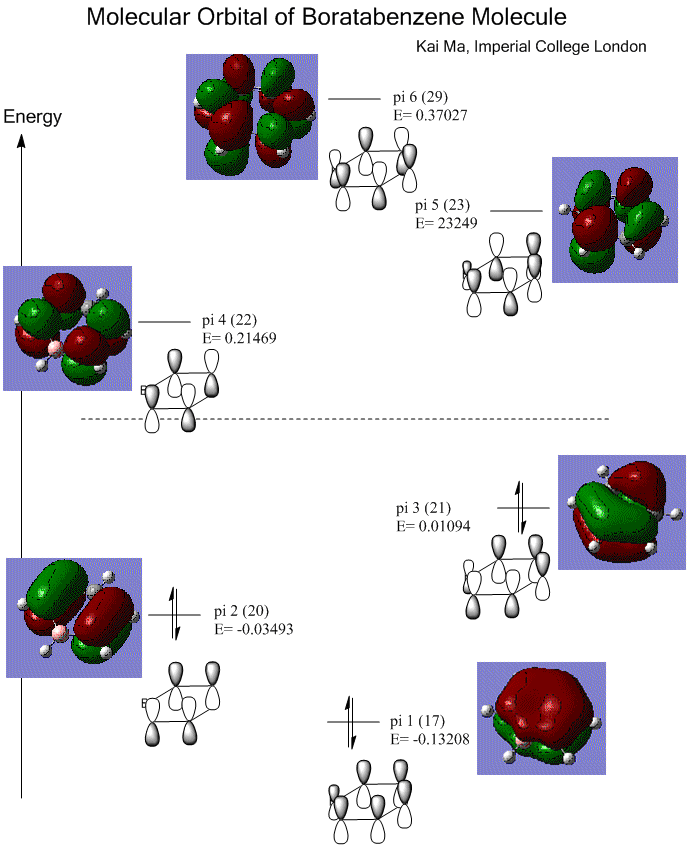
1. Molecular Orbital of BH3
Below is the predicted molecular orbital of BH3 molecule. The calculation in Gaussian involves in B3LYP method and 6-31G(d,p) basis set, as well as full population and full NBO.
The bond order = 1 accordingly.
2. Visualized Molecular Orbital of BH3
Here is the visualized occupied molecular orbitals of BH3 molecule in Gaussview.
| Visualized Orbital |  |
 |
 |
 |
 |
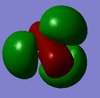 |
 |

|
| Symmetry Label | a1' | a1' | e' | e' | a2' | e' | e' | a1' |
3. Result and Discussion
The visualized molecular orbitals matches well with the predicted LCAO. The advantages of visualization is that it provides more clear and realistic model of how the electron cloud is distributed as in LCAO, we simply describe the combination of orbitals by overlapping the orbitals regardless of the distortion. It shows the HOMO and LUMO clearly in a 3D version and can be viewed in different angles.
However, a reordering of orbitals also occurs during visualization as the 3a1' orbital goes higher in energy and beyond the energy of 2e' orbital. It indicates that in cases of orbitals involving high frequencies, the visualization tends to be less accurate due to the limitation of method.
4. File Link
To access to the D-space link, click here.
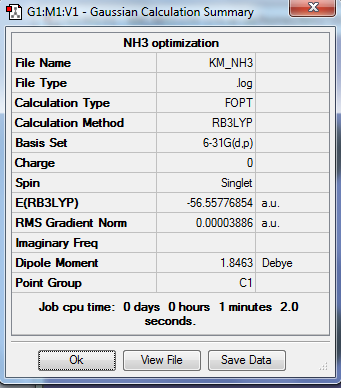
Analysis of NH3 Molecule
Optimization Analysis
B3LYP method and 6-31G(d,p) basis set (exactly the same as what has been done to BH3 molecule) was used in optimization of NH3 because it only involves in 4 atoms without any heavy atom.
1. Optimized NH3 Molecule
test molecule |
2. Geometry Data
Bond length for each B-H: 1.018 Å
Bond angel between each B-H: 105.7o
3. Calculation Summary
4. Real Output of Optimized NH3 molecule
Item Value Threshold Converged?
Maximum Force 0.000077 0.000450 YES
RMS Force 0.000044 0.000300 YES
Maximum Displacement 0.000174 0.001800 YES
RMS Displacement 0.000112 0.001200 YES
Predicted change in Energy=-1.601541D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = -0.0001 !
! R2 R(1,3) 1.018 -DE/DX = -0.0001 !
! R3 R(1,4) 1.0181 -DE/DX = -0.0001 !
! A1 A(2,1,3) 105.7546 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7438 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.743 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8739 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
5. File Link
To access to the file link, click here.
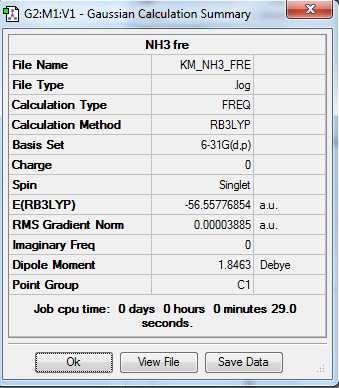
Frequency Analysis
This is carried out from the optimized molecule from previous session.
1. Calculation Summary
2. Real Output of Frequency Analysis of NH3
Low frequencies --- -9.0910 -0.0012 0.0005 0.0005 33.0170 39.6766
Low frequencies --- 1089.7840 1694.1623 1694.3189
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A A
Frequencies -- 1089.7840 1694.1623 1694.3189
Red. masses -- 1.1800 1.0645 1.0644
Frc consts -- 0.8257 1.8001 1.8004
IR Inten -- 145.4156 13.5550 13.5514
Atom AN X Y Z X Y Z X Y Z
1 7 0.12 0.00 0.00 0.00 0.02 -0.06 0.00 0.06 0.02
2 1 -0.53 -0.21 0.00 0.08 0.05 0.72 0.25 0.14 -0.24
3 1 -0.53 0.11 0.18 0.17 -0.54 0.20 -0.19 -0.38 0.35
4 1 -0.53 0.11 -0.18 -0.25 0.20 -0.05 -0.05 -0.63 -0.40
3. Vibration Modes of NH3
| No. | Vibration Mode | Description | Frequency/ cm-1 | Literature Frequency/ cm-1[5] | Intensity |
|---|---|---|---|---|---|
| 1 |  |
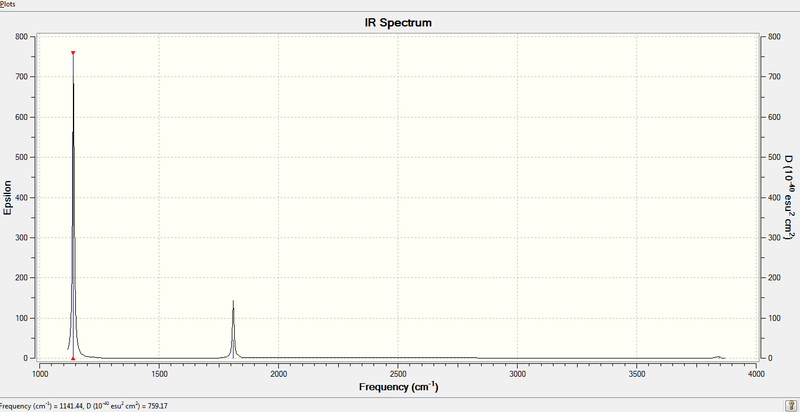
All H atoms move in a plane concertedly while N atom moves in the opposite direction. This leads to a large change in dipole moment. | 1089.78 | 1141 | 145.4156 |
| 2 |  |
Two H atoms rock together while the remaining H atom moves in the opposite direction. N atom moves slightly towards the single H atom. The resultant dipole moment is small. | 1694.16 | 1812 | 13.5550 |
| 3 |  |
Two H atoms stretch in and put in a concerted manner, towards the opposite direction of the other H atom. | 1694.32 | 1812 | 13.5514 |
| 4 |  |
All four H atoms stretch in and out concertedly. This results in a very slight change in dipole moment. | 3460.24 | 3706 | 1.0611 |
| 5 |  |
Two H atoms stretch at the opposite direction of the remaining one. N atom stays fixed in position and results in no change in dipole moment. This vibration mode is therefore not IR active. | 3588.59 | 3844 | 0.2729 |
| 6 |  |
One H atom stretches at the opposite direction of another H atom while the third H atom and N atom stays unchanged. Dipole moment is not changed in this mode. | 3588.84 | 3844 | 0.2702 |
4. IR Spectrum of NH3
5. File Link
To access to the file link, click here.
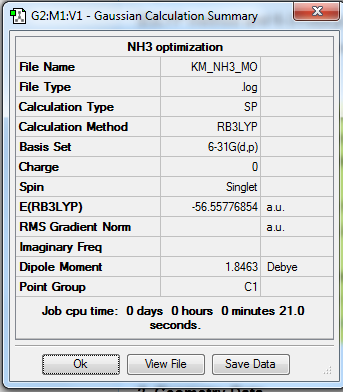
Molecular Orbital Analysis
B3LYP method and 6-31G(d,p) basis set is still used with full population and full NBOs.
To access the file link, click here.
NBO Analysis
1. Calculation Summary
2. Charge Distribution of NH3
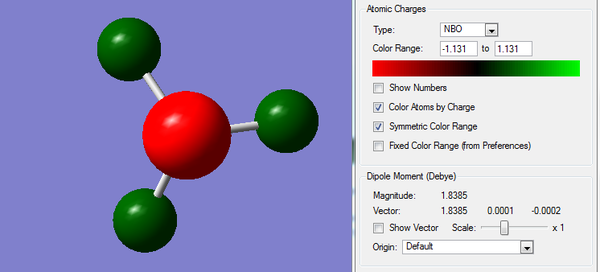
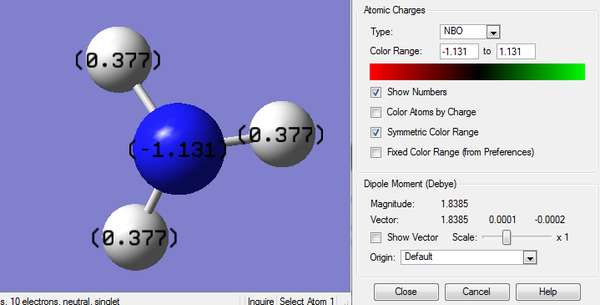
The calculation is carried out from previously optimized NH3 molecule. The color range is set from -1.131 to +1.131 as shown below.
The red region indicates highly negative charges, which is around the N atom and green region, indicating highly positive charges, is distributed around H atoms.
3. Charge Number of NH3
N atom has a very negative charge number due to the electronegative nature.
4. Real Output of NBO Analysis for NH3
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
N 1 -1.12518 1.99982 6.11108 0.01429 8.12518
H 2 0.37506 0.00000 0.62248 0.00246 0.62494
H 3 0.37506 0.00000 0.62248 0.00246 0.62494
H 4 0.37506 0.00000 0.62249 0.00246 0.62494
=======================================================================
* Total * 0.00000 1.99982 7.97853 0.02166 10.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.99909) BD ( 1) N 1 - H 2
( 68.83%) 0.8297* N 1 s( 24.87%)p 3.02( 75.05%)d 0.00( 0.09%)
-0.0001 -0.4986 -0.0059 0.0000 -0.2910
0.0052 0.8155 0.0277 0.0000 0.0000
0.0281 0.0000 0.0000 0.0032 0.0082
( 31.17%) 0.5583* H 2 s( 99.91%)p 0.00( 0.09%)
-0.9996 0.0000 0.0072 -0.0289 0.0000
2. (1.99909) BD ( 1) N 1 - H 3
( 68.83%) 0.8297* N 1 s( 24.87%)p 3.02( 75.05%)d 0.00( 0.09%)
0.0001 0.4986 0.0059 0.0000 0.2910
-0.0052 0.4078 0.0138 0.7062 0.0239
0.0140 0.0243 0.0076 0.0033 0.0031
( 31.17%) 0.5583* H 3 s( 99.91%)p 0.00( 0.09%)
0.9996 0.0000 -0.0072 -0.0145 -0.0250
3. (1.99909) BD ( 1) N 1 - H 4
( 68.83%) 0.8297* N 1 s( 24.86%)p 3.02( 75.05%)d 0.00( 0.09%)
0.0001 0.4986 0.0059 0.0000 0.2910
-0.0052 0.4078 0.0138 -0.7063 -0.0239
0.0140 -0.0243 -0.0076 0.0033 0.0031
( 31.17%) 0.5583* H 4 s( 99.91%)p 0.00( 0.09%)
0.9996 0.0000 -0.0072 -0.0145 0.0250
4. (1.99982) CR ( 1) N 1 s(100.00%)
1.0000 -0.0002 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
5. (1.99721) LP ( 1) N 1 s( 25.38%)p 2.94( 74.52%)d 0.00( 0.10%)
0.0001 0.5037 -0.0120 0.0000 -0.8618
0.0505 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 -0.0269 0.0155
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (H3N)
1. BD ( 1) N 1 - H 2 1.99909 -0.60415
2. BD ( 1) N 1 - H 3 1.99909 -0.60414
3. BD ( 1) N 1 - H 4 1.99909 -0.60411
4. CR ( 1) N 1 1.99982 -14.16772
5. LP ( 1) N 1 1.99721 -0.31757 16(v),20(v),24(v),25(v)
17(v),21(v)
Analysis of Ammonia-Borane Molecule
Optimization of Ammonia-Borane Molecule
1. Optimized Ammonia-Borane Molecule
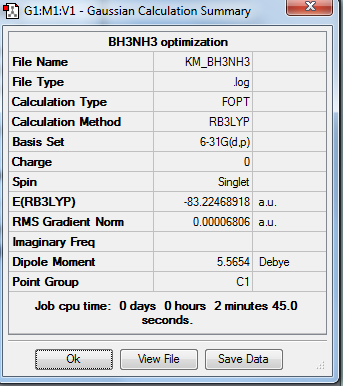
B3LYP method and 6-31G(d,p) basis set is used in optimization of this molecule and the results are shown as below.
test molecule |
2. Calculation Summary
3. Real Output of Optimized Ammonia-Borane Molecule
Item Value Threshold Converged?
Maximum Force 0.000137 0.000450 YES
RMS Force 0.000063 0.000300 YES
Maximum Displacement 0.000606 0.001800 YES
RMS Displacement 0.000336 0.001200 YES
Predicted change in Energy=-1.994007D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,7) 1.0186 -DE/DX = -0.0001 !
! R2 R(2,7) 1.0186 -DE/DX = -0.0001 !
! R3 R(3,7) 1.0186 -DE/DX = -0.0001 !
! R4 R(4,8) 1.2101 -DE/DX = -0.0001 !
! R5 R(5,8) 1.2101 -DE/DX = -0.0001 !
! R6 R(6,8) 1.2101 -DE/DX = -0.0001 !
! R7 R(7,8) 1.668 -DE/DX = -0.0001 !
! A1 A(1,7,2) 107.87 -DE/DX = 0.0 !
! A2 A(1,7,3) 107.8652 -DE/DX = 0.0 !
! A3 A(1,7,8) 111.0329 -DE/DX = 0.0 !
! A4 A(2,7,3) 107.8697 -DE/DX = 0.0 !
! A5 A(2,7,8) 111.0286 -DE/DX = 0.0 !
! A6 A(3,7,8) 111.0291 -DE/DX = 0.0 !
! A7 A(4,8,5) 113.8693 -DE/DX = 0.0 !
! A8 A(4,8,6) 113.8721 -DE/DX = 0.0 !
! A9 A(4,8,7) 104.6003 -DE/DX = 0.0 !
! A10 A(5,8,6) 113.8747 -DE/DX = 0.0 !
! A11 A(5,8,7) 104.6003 -DE/DX = 0.0 !
! A12 A(6,8,7) 104.5984 -DE/DX = 0.0 !
! D1 D(1,7,8,4) -179.9867 -DE/DX = 0.0 !
! D2 D(1,7,8,5) -59.9892 -DE/DX = 0.0 !
! D3 D(1,7,8,6) 60.0135 -DE/DX = 0.0 !
! D4 D(2,7,8,4) -59.9839 -DE/DX = 0.0 !
! D5 D(2,7,8,5) 60.0136 -DE/DX = 0.0 !
! D6 D(2,7,8,6) -179.9837 -DE/DX = 0.0 !
! D7 D(3,7,8,4) 60.0161 -DE/DX = 0.0 !
! D8 D(3,7,8,5) -179.9864 -DE/DX = 0.0 !
! D9 D(3,7,8,6) -59.9837 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
4. File Link
To access to the file link, click here.
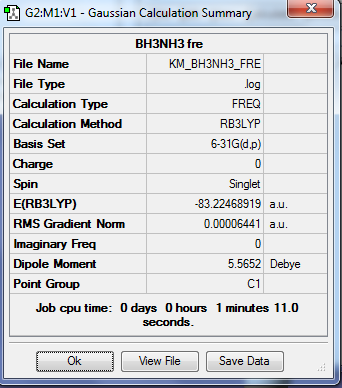
Frequency Analysis of Ammonia-Borane Molecule
Frequency analysis was carried our from previously optimized molecule.
1. Calculation Summary
2. Real Output of Frequency Analysis
Low frequencies --- -0.0015 -0.0011 0.0003 17.0498 22.5563 38.0324
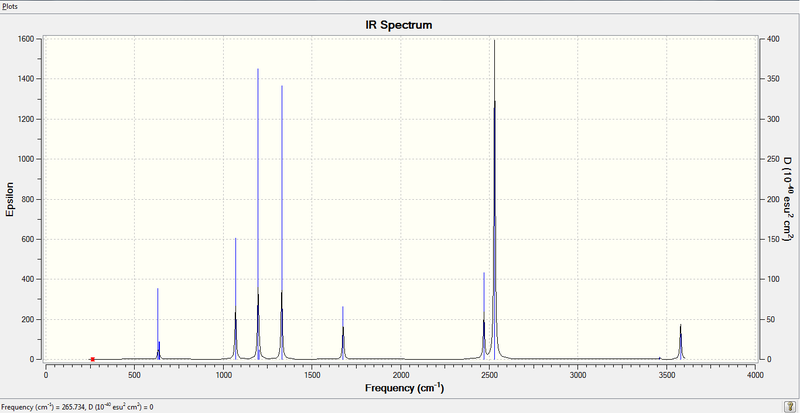
Low frequencies --- 265.7341 632.2424 638.9819
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A A
Frequencies -- 265.7336 632.2421 638.9819
Red. masses -- 1.0078 4.9962 1.0452
Frc consts -- 0.0419 1.1767 0.2514
IR Inten -- 0.0000 14.0050 3.5561
3. IR Spectrum of Ammonia-Borane Molecule
4. File Link
To access to the file link, click here.
Result and Discussion
1. Calculation of Association Energy of Ammonia-Borane Molecule
Balanced Equation: NH3 + BH3 -> H3B:NH3
E(NH3)= -56.55776854 a.u.
E(BH3)= -26.61532363 a.u.
E(NH3BH3)= -83.22468918 a.u.
ΔE = E(NH3BH3)-[E(NH3)+E(BH3)] = -83.22468918 - (-56.55776854 -26.61532363) = -0.05159701 a.u. = -135.4678447 kJ/mol-1
Comparing to literature, where the bond association energy is said to be -384.9 kJ/mol[6], there is a large difference. This might be resulted from the inaccuracy of the methods used in optimization. Also, the calculation in Gaussian assume that all the molecules are in vapor phase, which causes weakening of calculated bond strength.
2. Level of Accuracy
The error must be within a range of 10kJ/mol, which is 0.0038088 a.u.
Mini-Project: Investigating Aromaticity
In this mini project, aromaticity of different molecules is investigated via calculation and interpretation of the results. Calculation involves optimization which is carried out using 6-31G(d,p) basis set, frequency analysis, MO analysis and NBO analysis using full population. Based on the results obtained, some comparison and discussion has been made on the nature of aromaticity and the effect of replacing benzene with isoelectronic groups.
Benzene Molecule
Optimization of Benzene Molecule
1. Optimized Benzene Molecule Using 6-31G(d,p) Basis Set
test molecule |
2. Geometry Data
C-C Bond Length = 1.40 Å
C-H Bond Length = 1.09 Å
Bond Angle = 120o
3. Real Output of Optimized Benzene Molecule
Item Value Threshold Converged?
Maximum Force 0.000212 0.000450 YES
RMS Force 0.000085 0.000300 YES
Maximum Displacement 0.000991 0.001800 YES
RMS Displacement 0.000315 0.001200 YES
Predicted change in Energy=-5.157454D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3963 -DE/DX = 0.0001 !
! R2 R(1,6) 1.3961 -DE/DX = 0.0002 !
! R3 R(1,7) 1.0861 -DE/DX = 0.0002 !
! R4 R(2,3) 1.3961 -DE/DX = 0.0002 !
! R5 R(2,8) 1.0861 -DE/DX = 0.0002 !
! R6 R(3,4) 1.3963 -DE/DX = 0.0001 !
! R7 R(3,9) 1.086 -DE/DX = 0.0002 !
! R8 R(4,5) 1.3961 -DE/DX = 0.0002 !
! R9 R(4,10) 1.086 -DE/DX = 0.0002 !
! R10 R(5,6) 1.3963 -DE/DX = 0.0001 !
! R11 R(5,11) 1.0861 -DE/DX = 0.0002 !
! R12 R(6,12) 1.0861 -DE/DX = 0.0002 !
! A1 A(2,1,6) 119.9972 -DE/DX = 0.0 !
! A2 A(2,1,7) 119.9949 -DE/DX = 0.0 !
! A3 A(6,1,7) 120.0079 -DE/DX = 0.0 !
! A4 A(1,2,3) 120.0079 -DE/DX = 0.0 !
! A5 A(1,2,8) 119.9881 -DE/DX = 0.0 !
! A6 A(3,2,8) 120.004 -DE/DX = 0.0 !
! A7 A(2,3,4) 119.9948 -DE/DX = 0.0 !
! A8 A(2,3,9) 120.0086 -DE/DX = 0.0 !
! A9 A(4,3,9) 119.9966 -DE/DX = 0.0 !
! A10 A(3,4,5) 119.9972 -DE/DX = 0.0 !
! A11 A(3,4,10) 119.9934 -DE/DX = 0.0 !
! A12 A(5,4,10) 120.0094 -DE/DX = 0.0 !
! A13 A(4,5,6) 120.0083 -DE/DX = 0.0 !
! A14 A(4,5,11) 120.0014 -DE/DX = 0.0 !
! A15 A(6,5,11) 119.9904 -DE/DX = 0.0 !
! A16 A(1,6,5) 119.9946 -DE/DX = 0.0 !
! A17 A(1,6,12) 120.0106 -DE/DX = 0.0 !
! A18 A(5,6,12) 119.9948 -DE/DX = 0.0 !
! D1 D(6,1,2,3) -0.0059 -DE/DX = 0.0 !
! D2 D(6,1,2,8) 180.0023 -DE/DX = 0.0 !
! D3 D(7,1,2,3) -180.01 -DE/DX = 0.0 !
! D4 D(7,1,2,8) -0.0019 -DE/DX = 0.0 !
! D5 D(2,1,6,5) -0.0055 -DE/DX = 0.0 !
! D6 D(2,1,6,12) -179.9972 -DE/DX = 0.0 !
! D7 D(7,1,6,5) -180.0013 -DE/DX = 0.0 !
! D8 D(7,1,6,12) 0.007 -DE/DX = 0.0 !
! D9 D(1,2,3,4) 0.0117 -DE/DX = 0.0 !
! D10 D(1,2,3,9) -179.9914 -DE/DX = 0.0 !
! D11 D(8,2,3,4) 180.0036 -DE/DX = 0.0 !
! D12 D(8,2,3,9) 0.0005 -DE/DX = 0.0 !
! D13 D(2,3,4,5) -0.0062 -DE/DX = 0.0 !
! D14 D(2,3,4,10) -180.0059 -DE/DX = 0.0 !
! D15 D(9,3,4,5) 179.9969 -DE/DX = 0.0 !
! D16 D(9,3,4,10) -0.0028 -DE/DX = 0.0 !
! D17 D(3,4,5,6) -0.0051 -DE/DX = 0.0 !
! D18 D(3,4,5,11) 180.0058 -DE/DX = 0.0 !
! D19 D(10,4,5,6) -180.0055 -DE/DX = 0.0 !
! D20 D(10,4,5,11) 0.0054 -DE/DX = 0.0 !
! D21 D(4,5,6,1) 0.011 -DE/DX = 0.0 !
! D22 D(4,5,6,12) 180.0027 -DE/DX = 0.0 !
! D23 D(11,5,6,1) -179.9999 -DE/DX = 0.0 !
! D24 D(11,5,6,12) -0.0082 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
4. Calculation Summary
5. File Link
To access to the file of optimization, click here
Frequency Analysis of Benzene Molecule
1. Calculation Summary
2. Real Output of Frequency Analysis of Benzene Molecule
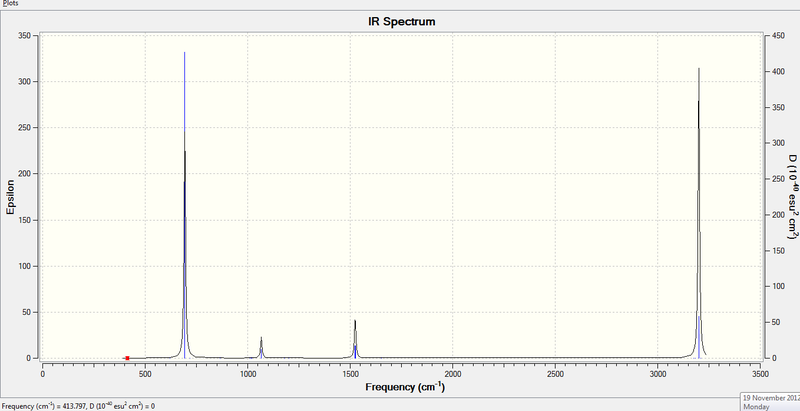
Low frequencies --- -17.2810 -14.5889 -9.6553 -0.0002 0.0007 0.0010
Low frequencies --- 413.7971 414.4696 620.8544
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A A
Frequencies -- 413.7971 414.4696 620.8539
Red. masses -- 2.9404 2.9434 6.0705
Frc consts -- 0.2966 0.2979 1.3786
IR Inten -- 0.0000 0.0000 0.0000
Atom AN X Y Z X Y Z X Y Z
1 6 0.00 0.00 -0.15 0.00 0.00 0.19 0.23 0.26 0.00
2 6 0.00 0.00 0.24 0.00 0.00 0.03 0.16 -0.05 0.00
3 6 0.00 0.00 -0.09 0.00 0.00 -0.22 0.22 -0.19 0.00
4 6 0.00 0.00 -0.15 0.00 0.00 0.19 -0.23 -0.26 0.00
5 6 0.00 0.00 0.24 0.00 0.00 0.03 -0.16 0.05 0.00
6 6 0.00 0.00 -0.09 0.00 0.00 -0.22 -0.22 0.19 0.00
7 1 0.00 0.00 -0.32 0.00 0.00 0.42 0.30 0.17 0.00
8 1 0.00 0.00 0.52 0.00 0.00 0.07 -0.20 -0.11 0.00
9 1 0.00 0.00 -0.20 0.00 0.00 -0.48 0.30 0.05 0.00
10 1 0.00 0.00 -0.32 0.00 0.00 0.42 -0.30 -0.17 0.00
11 1 0.00 0.00 0.52 0.00 0.00 0.07 0.20 0.11 0.00
12 1 0.00 0.00 -0.20 0.00 0.00 -0.48 -0.30 -0.05 0.00
3. IR Spectrum of Benzene Molecule
4. File Link
To access to file link, click here.
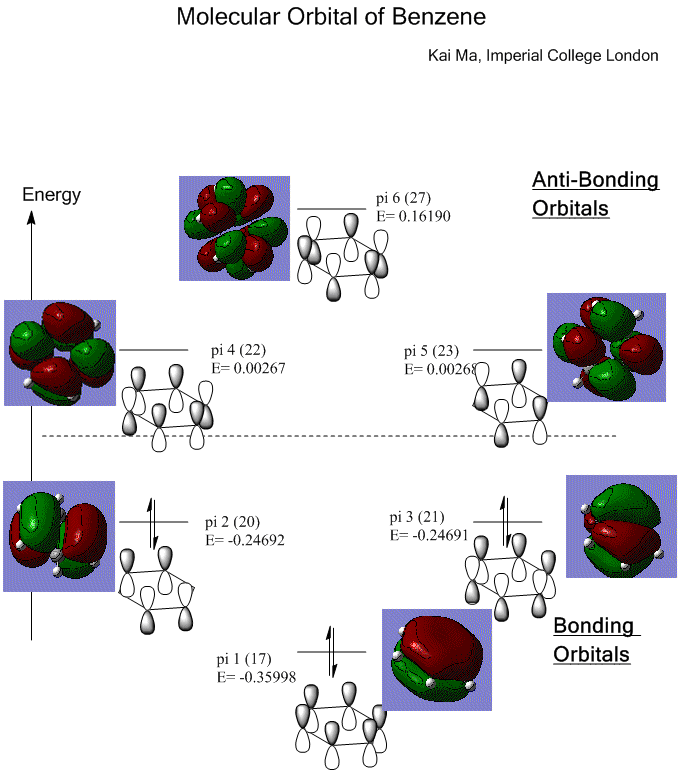
Molecular Orbital of Benzene Molecule
1. Predicted Molecular Orbital of Benzene
Benzene has a highly symmetrical structure, with all six carbons sp2 hybridized and identical, thus symmetric molecular orbitals shown as following.
Isovalue = 0.02.
Each of the carbon donates one pi electron, which in total, 6 pi electrons, occupy all the bonding orbitals. Therefore, benzene has an extremely stabilized structure.
Benzene is aromatic because it satisfies all the conditions according to Huckel’s rule:
Firstly, benzene has 4n+2 pi electrons where n=1 in this case;
Secondly, benzene has a closed ring structure;
Finally, benzene has a all its atoms in the same plane.
As shown in the molecular orbital, benzene also has highly electron- delocalized structure as the pi electrons distribute evenly along the carbon ring, which gives further stabilization of the total structure.
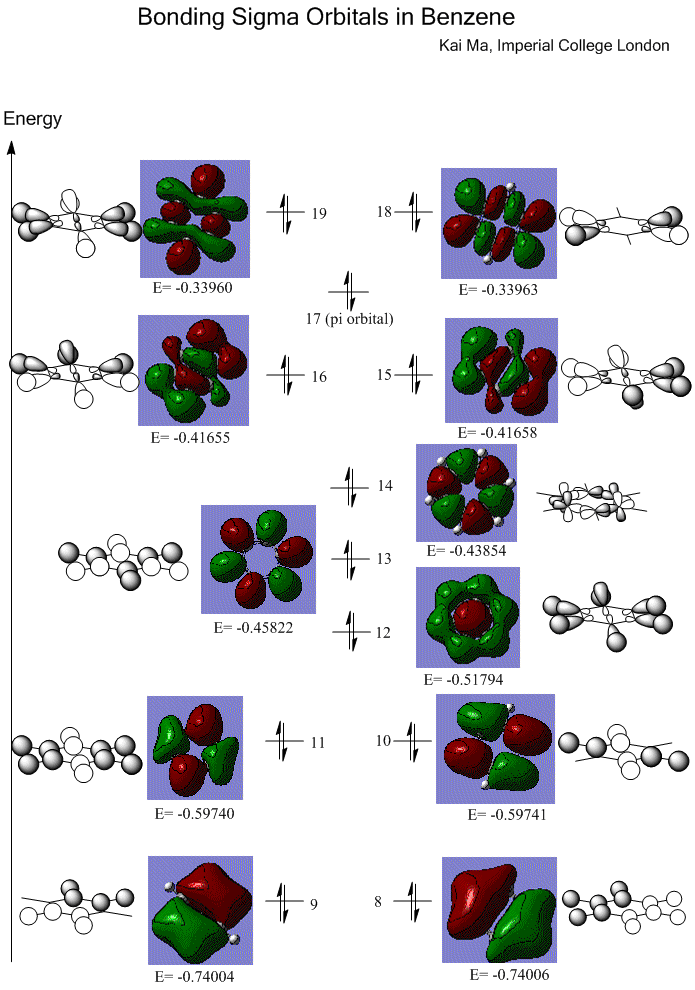
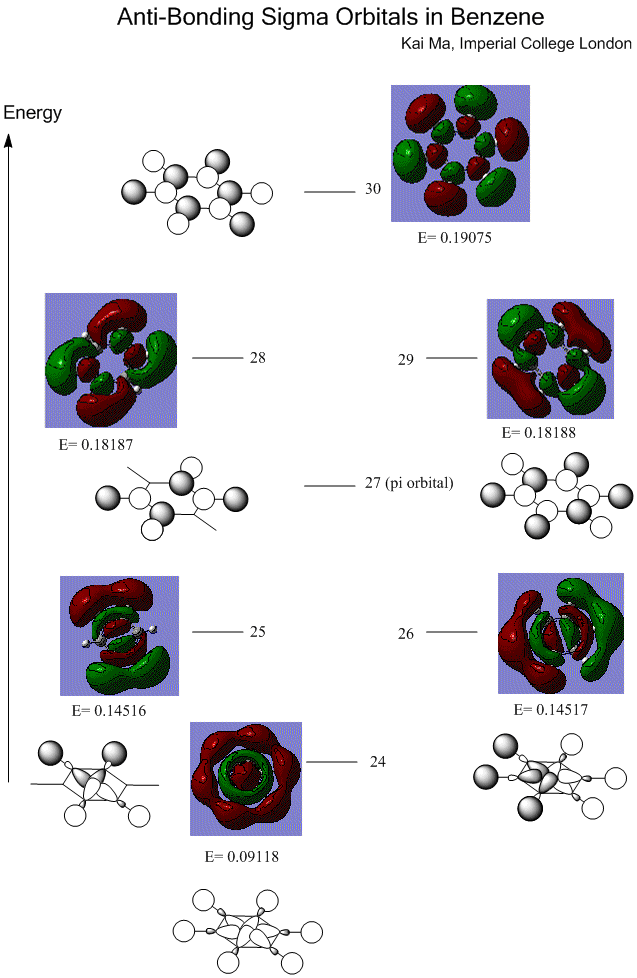
Sigma orbitals are also interpreted from the constructed MOs.
2. File Link
To access to the file link, click here.
NBO analysis of Benzene Molecule
1. Calculation Summary
2. Charge Distribution of Benzene Molecule
The analysis is carried out from previously optimized molecule with full population and full NBOs. The color range is set from -0.239 to +0.239 as shown below.
3. Charge Number of Benzene Molecule
4. Real Output of MO Analysis of Benzene Molecule
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
C 1 -0.23852 1.99910 4.22611 0.01331 6.23852
C 2 -0.23855 1.99910 4.22613 0.01331 6.23855
C 3 -0.23854 1.99910 4.22613 0.01331 6.23854
C 4 -0.23852 1.99910 4.22611 0.01331 6.23852
C 5 -0.23855 1.99910 4.22613 0.01331 6.23855
C 6 -0.23854 1.99910 4.22613 0.01331 6.23854
H 7 0.23854 0.00000 0.76003 0.00144 0.76146
H 8 0.23853 0.00000 0.76003 0.00144 0.76147
H 9 0.23854 0.00000 0.76002 0.00144 0.76146
H 10 0.23854 0.00000 0.76003 0.00144 0.76146
H 11 0.23853 0.00000 0.76003 0.00144 0.76147
H 12 0.23854 0.00000 0.76002 0.00144 0.76146
=======================================================================
* Total * 0.00000 11.99462 29.91690 0.08847 42.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98096) BD ( 1) C 1 - C 2
( 50.00%) 0.7071* C 1 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 0.7508
0.0048 -0.2875 -0.0354 0.0001 0.0000
-0.0092 0.0000 0.0000 0.0138 -0.0109
( 50.00%) 0.7071* C 2 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 -0.7645
-0.0260 0.2488 -0.0245 0.0000 0.0000
-0.0116 0.0000 0.0000 0.0119 -0.0109
2. (1.98097) BD ( 1) C 1 - C 6
( 50.00%) 0.7071* C 1 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 -0.1669
-0.0342 0.7864 0.0103 0.0000 0.0000
-0.0045 0.0000 0.0000 -0.0160 -0.0109
( 50.00%) 0.7071* C 6 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 0.1263
-0.0282 -0.7940 -0.0219 0.0000 0.0000
-0.0073 0.0000 0.0000 -0.0149 -0.0109
3. (1.66532) BD ( 2) C 1 - C 6
( 50.00%) 0.7071* C 1 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 -0.0001
0.0000 0.0000 0.0000 0.9997 -0.0133
0.0000 0.0066 0.0183 0.0000 0.0000
( 50.00%) 0.7071* C 6 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 -0.0001
0.0000 0.0000 0.0000 0.9997 -0.0133
0.0000 0.0128 -0.0147 0.0000 0.0000
4. (1.98305) BD ( 1) C 1 - H 7
( 62.04%) 0.7876* C 1 s( 29.58%)p 2.38( 70.39%)d 0.00( 0.04%)
0.0003 -0.5437 -0.0126 0.0010 0.6377
-0.0111 0.5450 -0.0095 0.0000 0.0000
-0.0164 0.0000 0.0000 -0.0026 0.0105
( 37.96%) 0.6161* H 7 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0014 -0.0173 -0.0148 0.0000
5. (1.98098) BD ( 1) C 2 - C 3
( 50.00%) 0.7071* C 2 s( 35.21%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 0.6244
0.0331 0.5064 -0.0135 0.0000 0.0000
0.0165 0.0000 0.0000 0.0011 -0.0109
( 50.00%) 0.7071* C 3 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 -0.5977
0.0082 -0.5377 -0.0348 0.0001 0.0000
0.0161 0.0000 0.0000 0.0041 -0.0109
6. (1.66534) BD ( 2) C 2 - C 3
( 50.00%) 0.7071* C 2 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9997 -0.0133
0.0000 0.0064 0.0184 0.0000 0.0000
( 50.00%) 0.7071* C 3 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 0.0001
0.0000 0.0000 0.0000 0.9997 -0.0133
0.0000 -0.0192 -0.0034 0.0000 0.0000
7. (1.98305) BD ( 1) C 2 - H 8
( 62.04%) 0.7876* C 2 s( 29.57%)p 2.38( 70.39%)d 0.00( 0.04%)
0.0003 -0.5436 -0.0126 0.0010 -0.1529
0.0027 0.8248 -0.0144 0.0000 0.0000
0.0060 0.0000 0.0000 0.0155 0.0105
( 37.96%) 0.6161* H 8 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0014 0.0042 -0.0224 0.0000
8. (1.98096) BD ( 1) C 3 - C 4
( 50.00%) 0.7071* C 3 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 -0.1264
0.0282 0.7940 0.0218 0.0000 0.0000
-0.0073 0.0000 0.0000 -0.0149 -0.0109
( 50.00%) 0.7071* C 4 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 0.1668
0.0342 -0.7865 -0.0103 0.0000 0.0000
-0.0045 0.0000 0.0000 -0.0160 -0.0109
9. (1.98305) BD ( 1) C 3 - H 9
( 62.04%) 0.7876* C 3 s( 29.58%)p 2.38( 70.38%)d 0.00( 0.04%)
-0.0003 0.5437 0.0126 -0.0010 0.7908
-0.0138 -0.2799 0.0049 -0.0001 0.0000
-0.0105 0.0000 0.0000 0.0129 -0.0105
( 37.96%) 0.6161* H 9 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0014 -0.0215 0.0076 0.0000
10. (1.98098) BD ( 1) C 4 - C 5
( 50.00%) 0.7071* C 4 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 -0.7508
-0.0048 0.2875 0.0354 0.0001 0.0000
-0.0092 0.0000 0.0000 0.0138 -0.0109
( 50.00%) 0.7071* C 5 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 0.7645
0.0260 -0.2487 0.0245 0.0000 0.0000
-0.0116 0.0000 0.0000 0.0119 -0.0109
11. (1.66533) BD ( 2) C 4 - C 5
( 50.00%) 0.7071* C 4 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 -0.0001 0.0000 0.9997 -0.0133
0.0000 -0.0191 -0.0037 0.0000 0.0000
( 50.00%) 0.7071* C 5 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9997 -0.0133
0.0000 0.0125 -0.0149 0.0000 0.0000
12. (1.98305) BD ( 1) C 4 - H 10
( 62.04%) 0.7876* C 4 s( 29.58%)p 2.38( 70.39%)d 0.00( 0.04%)
-0.0003 0.5437 0.0126 -0.0010 0.6378
-0.0111 0.5449 -0.0095 0.0000 0.0000
0.0164 0.0000 0.0000 0.0026 -0.0105
( 37.96%) 0.6161* H 10 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0014 -0.0173 -0.0148 0.0000
13. (1.98096) BD ( 1) C 5 - C 6
( 50.00%) 0.7071* C 5 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 -0.6244
-0.0330 -0.5064 0.0135 -0.0001 0.0000
0.0165 0.0000 0.0000 0.0011 -0.0109
( 50.00%) 0.7071* C 6 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 0.5978
-0.0082 0.5377 0.0347 0.0001 0.0000
0.0161 0.0000 0.0000 0.0041 -0.0109
14. (1.98305) BD ( 1) C 5 - H 11
( 62.04%) 0.7876* C 5 s( 29.57%)p 2.38( 70.39%)d 0.00( 0.04%)
-0.0003 0.5436 0.0126 -0.0010 -0.1531
0.0027 0.8248 -0.0143 0.0000 0.0000
-0.0060 0.0000 0.0000 -0.0155 -0.0105
( 37.96%) 0.6161* H 11 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0014 0.0042 -0.0224 0.0000
15. (1.98305) BD ( 1) C 6 - H 12
( 62.04%) 0.7876* C 6 s( 29.58%)p 2.38( 70.38%)d 0.00( 0.04%)
0.0003 -0.5437 -0.0126 0.0010 0.7907
-0.0138 -0.2800 0.0049 0.0000 0.0000
0.0105 0.0000 0.0000 -0.0129 0.0105
( 37.96%) 0.6161* H 12 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0014 -0.0215 0.0076 0.0000
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (C6H6)
1. BD ( 1) C 1 - C 2 1.98096 -0.68184 110(g),107(g),120(v),114(v)
43(v),73(v),109(g),112(g)
72(v),42(v)
2. BD ( 1) C 1 - C 6 1.98097 -0.68199 118(g),106(g),119(v),112(v)
63(v),33(v),120(g),109(g)
32(v),62(v)
3. BD ( 2) C 1 - C 6 1.66532 -0.23794 116(v),111(v),35(v),65(v)
4. BD ( 1) C 1 - H 7 1.98305 -0.51236 118(v),110(v),72(v),32(v)
107(g),106(g)
5. BD ( 1) C 2 - C 3 1.98098 -0.68202 106(g),113(g),117(v),109(v)
23(v),53(v),114(g),112(g)
52(v),22(v)
6. BD ( 2) C 2 - C 3 1.66534 -0.23795 116(v),108(v),25(v),55(v)
7. BD ( 1) C 2 - H 8 1.98305 -0.51233 113(v),107(v),42(v),22(v)
110(g),106(g)
8. BD ( 1) C 3 - C 4 1.98096 -0.68183 110(g),115(g),112(v),119(v)
33(v),63(v),114(g),117(g)
62(v),32(v)
9. BD ( 1) C 3 - H 9 1.98305 -0.51236 106(v),115(v),32(v),52(v)
110(g),113(g)
10. BD ( 1) C 4 - C 5 1.98098 -0.68200 118(g),113(g),114(v),120(v)
73(v),43(v),117(g),119(g)
42(v),72(v)
11. BD ( 2) C 4 - C 5 1.66533 -0.23794 108(v),111(v),45(v),75(v)
12. BD ( 1) C 4 - H 10 1.98305 -0.51236 118(v),110(v),62(v),42(v)
115(g),113(g)
13. BD ( 1) C 5 - C 6 1.98096 -0.68186 115(g),107(g),109(v),117(v)
53(v),23(v),119(g),120(g)
22(v),52(v)
14. BD ( 1) C 5 - H 11 1.98305 -0.51233 113(v),107(v),52(v),72(v)
115(g),118(g)
15. BD ( 1) C 6 - H 12 1.98305 -0.51236 106(v),115(v),22(v),62(v)
107(g),118(g)
16. CR ( 1) C 1 1.99911 -10.04057 73(v),33(v),110(v),118(v)
120(v),112(v)
17. CR ( 1) C 2 1.99911 -10.04056 43(v),23(v),113(v),107(v)
114(v),109(v)
18. CR ( 1) C 3 1.99911 -10.04056 33(v),53(v),106(v),115(v)
112(v),117(v)
19. CR ( 1) C 4 1.99911 -10.04057 63(v),43(v),118(v),110(v)
119(v),114(v)
20. CR ( 1) C 5 1.99911 -10.04056 53(v),73(v),113(v),107(v)
117(v),120(v)
21. CR ( 1) C 6 1.99911 -10.04056 23(v),63(v),115(v),106(v)
109(v),119(v)
5. File Link
To access to the file link, click here.
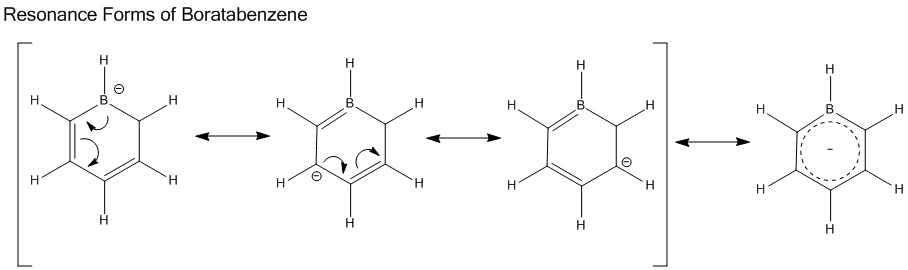
Analysis of Boratabenzene Molecule
Optimization of Boratabenzene Molecule
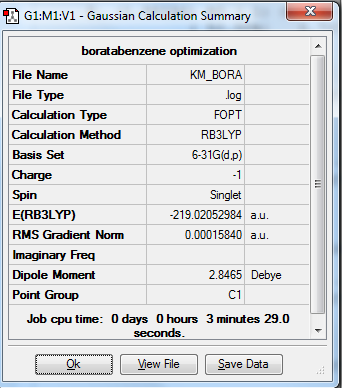
1. Optimized Boratabenzene Molecule using 6-31G(d,p) Basis Set
Boratabenzene molecule is optimized using B3LYP method and 6-31G(d,p) basis set.
test molecule |
2. Geometry Data
C-B Bond length = 1.51 Å
C-C Bond length = 1.40 Å
C-H Bond length = 1.10 Å
B-H Bond length = 1.21 Å
Bond angle = 120o
3. Real Output of Optimized Boratabenzene Molecule
Item Value Threshold Converged?
Maximum Force 0.000159 0.000450 YES
RMS Force 0.000069 0.000300 YES
Maximum Displacement 0.000878 0.001800 YES
RMS Displacement 0.000326 0.001200 YES
Predicted change in Energy=-6.589451D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.4053 -DE/DX = -0.0001 !
! R2 R(1,5) 1.3989 -DE/DX = 0.0 !
! R3 R(1,6) 1.0968 -DE/DX = 0.0001 !
! R4 R(2,3) 1.4053 -DE/DX = -0.0001 !
! R5 R(2,7) 1.0916 -DE/DX = -0.0001 !
! R6 R(3,4) 1.3989 -DE/DX = 0.0 !
! R7 R(3,8) 1.0968 -DE/DX = 0.0001 !
! R8 R(4,9) 1.097 -DE/DX = -0.0001 !
! R9 R(4,12) 1.5137 -DE/DX = 0.0001 !
! R10 R(5,11) 1.097 -DE/DX = -0.0001 !
! R11 R(5,12) 1.5138 -DE/DX = 0.0001 !
! R12 R(10,12) 1.2185 -DE/DX = 0.0 !
! A1 A(2,1,5) 122.138 -DE/DX = 0.0001 !
! A2 A(2,1,6) 117.4354 -DE/DX = 0.0 !
! A3 A(5,1,6) 120.4266 -DE/DX = -0.0002 !
! A4 A(1,2,3) 120.4508 -DE/DX = -0.0001 !
! A5 A(1,2,7) 119.7734 -DE/DX = 0.0001 !
! A6 A(3,2,7) 119.7758 -DE/DX = 0.0001 !
! A7 A(2,3,4) 122.1395 -DE/DX = 0.0001 !
! A8 A(2,3,8) 117.4371 -DE/DX = 0.0 !
! A9 A(4,3,8) 120.4234 -DE/DX = -0.0002 !
! A10 A(3,4,9) 115.9493 -DE/DX = 0.0001 !
! A11 A(3,4,12) 120.0806 -DE/DX = -0.0001 !
! A12 A(9,4,12) 123.9701 -DE/DX = -0.0001 !
! A13 A(1,5,11) 115.9535 -DE/DX = 0.0001 !
! A14 A(1,5,12) 120.0812 -DE/DX = -0.0001 !
! A15 A(11,5,12) 123.9654 -DE/DX = -0.0001 !
! A16 A(4,12,5) 115.1098 -DE/DX = 0.0 !
! A17 A(4,12,10) 122.4482 -DE/DX = 0.0 !
! A18 A(5,12,10) 122.4419 -DE/DX = 0.0 !
! D1 D(5,1,2,3) 0.0057 -DE/DX = 0.0 !
! D2 D(5,1,2,7) 180.0027 -DE/DX = 0.0 !
! D3 D(6,1,2,3) 180.0038 -DE/DX = 0.0 !
! D4 D(6,1,2,7) 0.0008 -DE/DX = 0.0 !
! D5 D(2,1,5,11) -180.0018 -DE/DX = 0.0 !
! D6 D(2,1,5,12) -0.001 -DE/DX = 0.0 !
! D7 D(6,1,5,11) 0.0002 -DE/DX = 0.0 !
! D8 D(6,1,5,12) 180.001 -DE/DX = 0.0 !
! D9 D(1,2,3,4) -0.0074 -DE/DX = 0.0 !
! D10 D(1,2,3,8) -180.0016 -DE/DX = 0.0 !
! D11 D(7,2,3,4) -180.0044 -DE/DX = 0.0 !
! D12 D(7,2,3,8) 0.0014 -DE/DX = 0.0 !
! D13 D(2,3,4,9) 180.0049 -DE/DX = 0.0 !
! D14 D(2,3,4,12) 0.0042 -DE/DX = 0.0 !
! D15 D(8,3,4,9) -0.0011 -DE/DX = 0.0 !
! D16 D(8,3,4,12) -180.0018 -DE/DX = 0.0 !
! D17 D(3,4,12,5) 0.0005 -DE/DX = 0.0 !
! D18 D(3,4,12,10) -180.0 -DE/DX = 0.0 !
! D19 D(9,4,12,5) -180.0003 -DE/DX = 0.0 !
! D20 D(9,4,12,10) -0.0008 -DE/DX = 0.0 !
! D21 D(1,5,12,4) -0.002 -DE/DX = 0.0 !
! D22 D(1,5,12,10) -180.0015 -DE/DX = 0.0 !
! D23 D(11,5,12,4) 179.9989 -DE/DX = 0.0 !
! D24 D(11,5,12,10) -0.0006 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
4. Calculation Summary
5. File Link
To access to file of optimization, click here.
Frequency Analysis of Boratabenzene Molecule
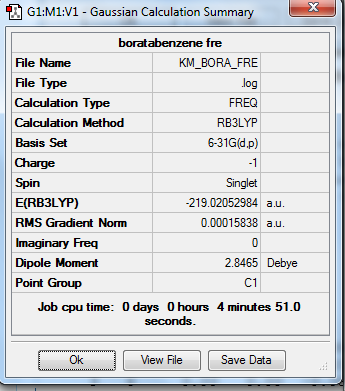
1. Calculation Summary
2. Real Output of Frequency Analysis of Boratabenzene Molecule
Low frequencies --- -13.1275 0.0003 0.0006 0.0008 15.0447 18.1653
Low frequencies --- 371.3454 404.2334 565.2534
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A A
Frequencies -- 371.3453 404.2327 565.2534
Red. masses -- 2.6885 3.2177 5.7648
Frc consts -- 0.2184 0.3098 1.0852
IR Inten -- 2.3000 0.0000 0.1576
Atom AN X Y Z X Y Z X Y Z
1 6 0.00 0.00 -0.08 0.00 0.00 0.22 0.23 0.21 0.00
2 6 0.00 0.00 0.21 0.00 0.00 0.00 0.14 0.00 0.00
3 6 0.00 0.00 -0.08 0.00 0.00 -0.22 0.23 -0.22 0.00
4 6 0.00 0.00 -0.14 0.00 0.00 0.23 -0.22 -0.21 0.00
5 6 0.00 0.00 -0.14 0.00 0.00 -0.23 -0.22 0.21 0.00
6 1 0.00 0.00 -0.20 0.00 0.00 0.52 0.31 0.08 0.00
7 1 0.00 0.00 0.38 0.00 0.00 0.00 -0.21 0.00 0.00
8 1 0.00 0.00 -0.20 0.00 0.00 -0.52 0.31 -0.08 0.00
9 1 0.00 0.00 -0.35 0.00 0.00 0.36 -0.34 0.06 0.00
10 1 0.00 0.00 0.62 0.00 0.00 0.00 0.29 0.00 0.00
11 1 0.00 0.00 -0.35 0.00 0.00 -0.36 -0.34 -0.06 0.00
12 5 0.00 0.00 0.25 0.00 0.00 0.00 -0.17 0.00 0.00
3. IR Spectrum of Boratabenzene Molecule
4. File Link
To access to file link, click here.
Molecular Orbital of Boratabenzene Molecule
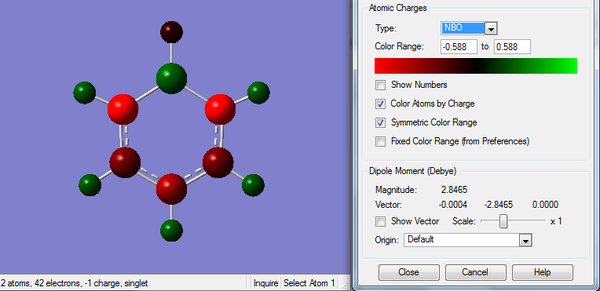
Since B is less electronegative than C atom, electron density is pulled away from B atom. It is also less symmetric than benzene due to the replacement of B-H unit.
Isovalue = 0.03.
NBO analysis of Boratabenzene Molecule
1. Calculation Summary
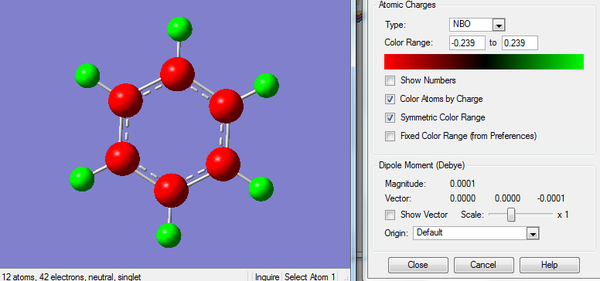
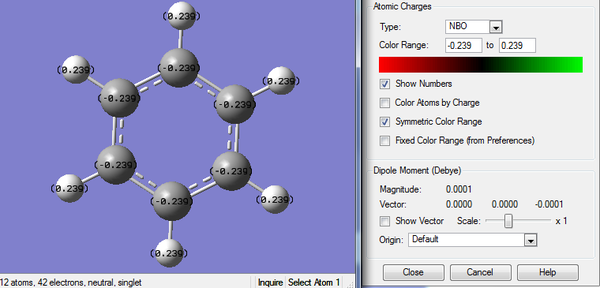
2. Charge Distribution of Boratabenzene Molecule
The color range is set to -0.588 to +0.588.
3. Charge Number of Boratabenzene Molecule
4. Real Output of MO Analysis of Boratabenzene Molecule
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
C 1 -0.25043 1.99910 4.23720 0.01412 6.25043
C 2 -0.33984 1.99907 4.32693 0.01384 6.33984
C 3 -0.25041 1.99910 4.23719 0.01412 6.25041
C 4 -0.58795 1.99901 4.57715 0.01178 6.58795
C 5 -0.58790 1.99901 4.57711 0.01178 6.58790
H 6 0.17906 0.00000 0.81832 0.00262 0.82094
H 7 0.18564 0.00000 0.81237 0.00200 0.81436
H 8 0.17906 0.00000 0.81832 0.00262 0.82094
H 9 0.18380 0.00000 0.81402 0.00218 0.81620
H 10 -0.09642 0.00000 1.09588 0.00054 1.09642
H 11 0.18380 0.00000 0.81402 0.00218 0.81620
B 12 0.20160 1.99906 2.78774 0.01160 4.79840
=======================================================================
* Total * -1.00000 11.99436 29.91625 0.08939 42.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.97970) BD ( 1) C 1 - C 2
( 49.96%) 0.7068* C 1 s( 35.50%)p 1.82( 64.46%)d 0.00( 0.04%)
-0.0001 0.5958 -0.0075 0.0006 0.6874
0.0034 -0.4135 -0.0325 0.0000 0.0000
-0.0146 0.0000 0.0000 0.0081 -0.0107
( 50.04%) 0.7074* C 2 s( 35.88%)p 1.79( 64.09%)d 0.00( 0.04%)
-0.0001 0.5989 -0.0072 0.0010 -0.7062
-0.0327 0.3754 -0.0141 0.0000 0.0000
-0.0137 0.0000 0.0000 0.0078 -0.0107
2. (1.98270) BD ( 1) C 1 - C 5
( 50.77%) 0.7125* C 1 s( 37.60%)p 1.66( 62.37%)d 0.00( 0.03%)
-0.0001 0.6131 -0.0079 0.0007 -0.0573
-0.0311 0.7869 0.0164 0.0000 0.0000
-0.0020 0.0000 0.0000 -0.0150 -0.0098
( 49.23%) 0.7017* C 5 s( 32.49%)p 2.08( 67.46%)d 0.00( 0.05%)
0.0000 0.5697 -0.0200 0.0010 0.0025
-0.0269 -0.8201 -0.0353 0.0000 0.0000
0.0006 0.0000 0.0000 -0.0173 -0.0123
3. (1.76867) BD ( 2) C 1 - C 5
( 48.13%) 0.6938* C 1 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9996 -0.0213
0.0000 0.0031 0.0171 0.0000 0.0000
( 51.87%) 0.7202* C 5 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9998 -0.0054
0.0000 0.0016 -0.0185 0.0000 0.0000
4. (1.98570) BD ( 1) C 1 - H 6
( 59.32%) 0.7702* C 1 s( 26.88%)p 2.72( 73.08%)d 0.00( 0.05%)
0.0003 -0.5182 -0.0133 0.0012 0.7228
-0.0089 0.4562 -0.0100 0.0000 0.0000
-0.0177 0.0000 0.0000 -0.0069 0.0111
( 40.68%) 0.6378* H 6 s( 99.95%)p 0.00( 0.05%)
-0.9998 -0.0026 -0.0187 -0.0116 0.0000
5. (1.97971) BD ( 1) C 2 - C 3
( 50.04%) 0.7074* C 2 s( 35.88%)p 1.79( 64.09%)d 0.00( 0.04%)
-0.0001 0.5989 -0.0072 0.0010 0.7063
0.0327 0.3752 -0.0141 0.0000 0.0000
0.0137 0.0000 0.0000 0.0078 -0.0107
( 49.96%) 0.7068* C 3 s( 35.51%)p 1.82( 64.45%)d 0.00( 0.04%)
-0.0001 0.5958 -0.0075 0.0006 -0.6875
-0.0034 -0.4133 -0.0325 0.0000 0.0000
0.0146 0.0000 0.0000 0.0081 -0.0107
6. (1.98507) BD ( 1) C 2 - H 7
( 59.44%) 0.7710* C 2 s( 28.22%)p 2.54( 71.74%)d 0.00( 0.04%)
0.0004 -0.5311 -0.0116 0.0020 0.0001
0.0000 0.8469 -0.0076 0.0000 0.0000
0.0000 0.0000 0.0000 0.0178 0.0110
( 40.56%) 0.6369* H 7 s( 99.95%)p 0.00( 0.05%)
-0.9998 -0.0011 0.0000 -0.0217 0.0000
7. (1.98270) BD ( 1) C 3 - C 4
( 50.77%) 0.7125* C 3 s( 37.60%)p 1.66( 62.37%)d 0.00( 0.03%)
-0.0001 0.6131 -0.0079 0.0007 0.0575
0.0311 0.7869 0.0164 0.0000 0.0000
0.0020 0.0000 0.0000 -0.0150 -0.0098
( 49.23%) 0.7017* C 4 s( 32.49%)p 2.08( 67.46%)d 0.00( 0.05%)
0.0000 0.5697 -0.0200 0.0010 -0.0027
0.0269 -0.8202 -0.0353 0.0000 0.0000
-0.0006 0.0000 0.0000 -0.0173 -0.0123
8. (1.76862) BD ( 2) C 3 - C 4
( 48.13%) 0.6937* C 3 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9996 -0.0213
0.0000 -0.0031 0.0171 0.0000 0.0000
( 51.87%) 0.7202* C 4 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9998 -0.0054
0.0000 -0.0016 -0.0185 0.0000 0.0000
9. (1.98570) BD ( 1) C 3 - H 8
( 59.32%) 0.7702* C 3 s( 26.88%)p 2.72( 73.08%)d 0.00( 0.05%)
-0.0003 0.5182 0.0133 -0.0012 0.7227
-0.0089 -0.4563 0.0100 0.0000 0.0000
-0.0177 0.0000 0.0000 0.0069 -0.0111
( 40.68%) 0.6378* H 8 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0026 -0.0187 0.0116 0.0000
10. (1.98420) BD ( 1) C 4 - H 9
( 59.41%) 0.7708* C 4 s( 25.38%)p 2.94( 74.57%)d 0.00( 0.05%)
-0.0003 0.5038 -0.0051 -0.0025 0.7907
-0.0003 0.3469 0.0088 0.0000 0.0000
0.0111 0.0000 0.0000 0.0150 -0.0119
( 40.59%) 0.6371* H 9 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0005 -0.0192 -0.0100 0.0000
11. (1.96997) BD ( 1) C 4 - B 12
( 66.70%) 0.8167* C 4 s( 42.04%)p 1.38( 57.96%)d 0.00( 0.01%)
0.0000 -0.6482 -0.0158 -0.0012 0.6115
-0.0293 -0.4524 -0.0090 0.0000 0.0000
0.0059 0.0000 0.0000 -0.0041 0.0057
( 33.30%) 0.5771* B 12 s( 33.40%)p 1.99( 66.52%)d 0.00( 0.08%)
0.0000 -0.5779 0.0059 -0.0048 -0.7056
-0.0393 0.4071 -0.0096 0.0000 0.0000
0.0230 0.0000 0.0000 -0.0082 0.0133
12. (1.98420) BD ( 1) C 5 - H 11
( 59.41%) 0.7708* C 5 s( 25.39%)p 2.94( 74.57%)d 0.00( 0.05%)
-0.0003 0.5038 -0.0051 -0.0025 -0.7906
0.0003 0.3471 0.0088 0.0000 0.0000
-0.0111 0.0000 0.0000 0.0149 -0.0119
( 40.59%) 0.6371* H 11 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0005 0.0192 -0.0100 0.0000
13. (1.96996) BD ( 1) C 5 - B 12
( 66.70%) 0.8167* C 5 s( 42.03%)p 1.38( 57.96%)d 0.00( 0.01%)
0.0000 0.6481 0.0158 0.0012 0.6117
-0.0293 0.4522 0.0090 0.0000 0.0000
0.0059 0.0000 0.0000 0.0041 -0.0057
( 33.30%) 0.5771* B 12 s( 33.40%)p 1.99( 66.52%)d 0.00( 0.08%)
0.0000 0.5779 -0.0059 0.0048 -0.7057
-0.0393 -0.4070 0.0096 0.0000 0.0000
0.0230 0.0000 0.0000 0.0082 -0.0133
14. (1.98604) BD ( 1) H 10 - B 12
( 55.09%) 0.7422* H 10 s( 99.97%)p 0.00( 0.03%)
0.9998 0.0001 0.0000 -0.0180 0.0000
( 44.91%) 0.6702* B 12 s( 33.16%)p 2.01( 66.78%)d 0.00( 0.06%)
-0.0005 0.5758 0.0069 -0.0060 0.0001
0.0000 0.8172 -0.0016 0.0000 0.0000
0.0000 0.0000 0.0000 -0.0213 -0.0105
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (C5H6B)
1. BD ( 1) C 1 - C 2 1.97970 -0.46973 108(g),111(g),115(v),118(v)
44(v),63(v),112(g),110(g)
2. BD ( 1) C 1 - C 5 1.98270 -0.46498 119(g),107(g),112(v),110(g)
34(v),120(v),118(g),98(v)
33(v)
3. BD ( 2) C 1 - C 5 1.76867 -0.02908 21(v),22(v),35(v),100(v)
109(g)
4. BD ( 1) C 1 - H 6 1.98570 -0.31413 111(v),119(v),33(v),63(v)
108(g)
5. BD ( 1) C 2 - C 3 1.97971 -0.46975 113(g),107(g),110(v),116(v)
24(v),53(v),112(g),115(g)
6. BD ( 1) C 2 - H 7 1.98507 -0.31744 113(v),108(v),43(v),23(v)
111(g),107(g)
7. BD ( 1) C 3 - C 4 1.98270 -0.46495 117(g),111(g),112(v),115(g)
34(v),120(v),116(g),98(v)
33(v)
8. BD ( 2) C 3 - C 4 1.76862 -0.02907 21(v),22(v),35(v),100(v)
114(g)
9. BD ( 1) C 3 - H 8 1.98570 -0.31412 107(v),117(v),33(v),53(v)
113(g)
10. BD ( 1) C 4 - H 9 1.98420 -0.28848 111(v),117(g),43(v),119(v)
97(v),113(g)
11. BD ( 1) C 4 - B 12 1.96997 -0.31779 113(g),115(v),118(v),116(g)
44(v),43(v),85(v),64(v)
119(g)
12. BD ( 1) C 5 - H 11 1.98420 -0.28849 107(v),119(g),23(v),117(v)
97(v),108(g)
13. BD ( 1) C 5 - B 12 1.96996 -0.31776 108(g),110(v),116(v),118(g)
24(v),23(v),93(v),54(v)
117(g)
14. BD ( 1) H 10 - B 12 1.98604 -0.17253 113(v),108(v),53(v),63(v)
15. CR ( 1) C 1 1.99910 -9.83478 64(v),34(v),119(v),112(v)
111(v),118(v)
16. CR ( 1) C 2 1.99907 -9.82827 44(v),24(v),113(v),108(v)
115(v),110(v),27(v),47(v)
43(v),23(v)
17. CR ( 1) C 3 1.99910 -9.83478 54(v),34(v),117(v),112(v)
107(v),116(v)
18. CR ( 1) C 4 1.99902 -9.79407 44(v),98(v),117(g),111(v)
115(v),97(v)
19. CR ( 1) C 5 1.99902 -9.79408 24(v),98(v),119(g),107(v)
110(v),97(v)
20. CR ( 1) B 12 1.99907 -6.36942 116(v),118(v),113(v),108(v)
53(v),63(v)
21. LP ( 1) C 2 1.14688 0.09689 114(v),109(v),35(g),46(v)
26(v),45(v),25(v)
22. LP*( 1) B 12 0.57262 0.22267 114(v),109(v),100(g),57(v)
5. File Link
To access to file link, click here
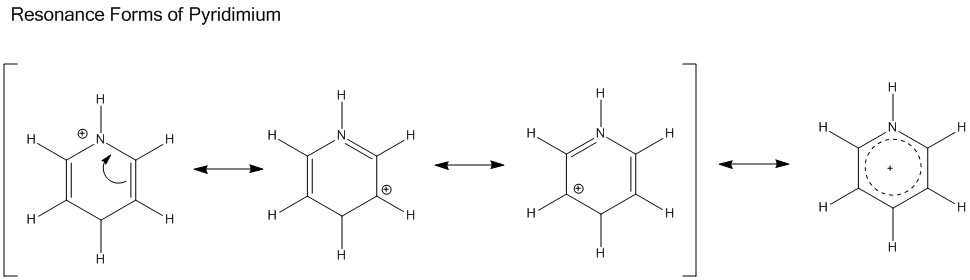
Analysis of Pyridinium Molecule
Optimization of Pyridinium Molecule
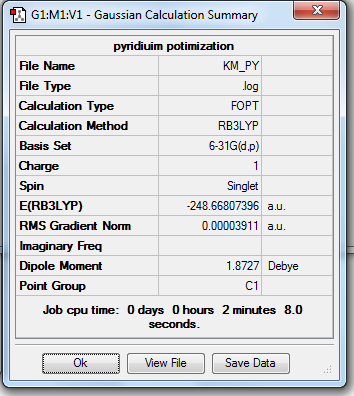
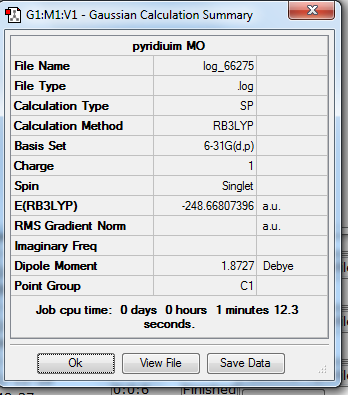
1. Optimized Pyridinium Molecule using 6-31G(d,p) Basis Set
test molecule |
2. Geometry Data
C-C Bond Length = 1.40 Å
C-N Bond Length = 1.35 Å
C-H Bond Length = 1.09 Å
N-H Bond Length = 1.01 Å
Bond Angle = 120o
3. Real Output of Optimized Pyridinium Molecule
Item Value Threshold Converged?
Maximum Force 0.000064 0.000450 YES
RMS Force 0.000023 0.000300 YES
Maximum Displacement 0.000822 0.001800 YES
RMS Displacement 0.000175 0.001200 YES
Predicted change in Energy=-6.915416D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3988 -DE/DX = 0.0 !
! R2 R(1,5) 1.3988 -DE/DX = 0.0 !
! R3 R(1,6) 1.0852 -DE/DX = 0.0 !
! R4 R(2,3) 1.3838 -DE/DX = 0.0 !
! R5 R(2,7) 1.0835 -DE/DX = 0.0 !
! R6 R(3,8) 1.0832 -DE/DX = 0.0 !
! R7 R(3,12) 1.3524 -DE/DX = 0.0 !
! R8 R(4,5) 1.3839 -DE/DX = 0.0 !
! R9 R(4,10) 1.0832 -DE/DX = 0.0 !
! R10 R(4,12) 1.3523 -DE/DX = 0.0001 !
! R11 R(5,11) 1.0835 -DE/DX = 0.0 !
! R12 R(9,12) 1.0169 -DE/DX = 0.0 !
! A1 A(2,1,5) 120.0549 -DE/DX = 0.0 !
! A2 A(2,1,6) 119.9711 -DE/DX = 0.0 !
! A3 A(5,1,6) 119.974 -DE/DX = 0.0 !
! A4 A(1,2,3) 119.0827 -DE/DX = 0.0 !
! A5 A(1,2,7) 121.4959 -DE/DX = -0.0001 !
! A6 A(3,2,7) 119.4215 -DE/DX = 0.0 !
! A7 A(2,3,8) 123.9326 -DE/DX = 0.0 !
! A8 A(2,3,12) 119.2355 -DE/DX = 0.0 !
! A9 A(8,3,12) 116.832 -DE/DX = 0.0 !
! A10 A(5,4,10) 123.9293 -DE/DX = 0.0 !
! A11 A(5,4,12) 119.2363 -DE/DX = 0.0 !
! A12 A(10,4,12) 116.8344 -DE/DX = 0.0 !
! A13 A(1,5,4) 119.082 -DE/DX = 0.0 !
! A14 A(1,5,11) 121.4987 -DE/DX = -0.0001 !
! A15 A(4,5,11) 119.4193 -DE/DX = 0.0001 !
! A16 A(3,12,4) 123.3087 -DE/DX = 0.0 !
! A17 A(3,12,9) 118.345 -DE/DX = 0.0 !
! A18 A(4,12,9) 118.3463 -DE/DX = 0.0 !
! D1 D(5,1,2,3) 0.0007 -DE/DX = 0.0 !
! D2 D(5,1,2,7) -180.0001 -DE/DX = 0.0 !
! D3 D(6,1,2,3) 180.0012 -DE/DX = 0.0 !
! D4 D(6,1,2,7) 0.0004 -DE/DX = 0.0 !
! D5 D(2,1,5,4) 0.0011 -DE/DX = 0.0 !
! D6 D(2,1,5,11) 180.0004 -DE/DX = 0.0 !
! D7 D(6,1,5,4) 180.0006 -DE/DX = 0.0 !
! D8 D(6,1,5,11) -0.0001 -DE/DX = 0.0 !
! D9 D(1,2,3,8) 179.9997 -DE/DX = 0.0 !
! D10 D(1,2,3,12) -0.0019 -DE/DX = 0.0 !
! D11 D(7,2,3,8) 0.0006 -DE/DX = 0.0 !
! D12 D(7,2,3,12) -180.0011 -DE/DX = 0.0 !
! D13 D(2,3,12,4) 0.0013 -DE/DX = 0.0 !
! D14 D(2,3,12,9) 180.0011 -DE/DX = 0.0 !
! D15 D(8,3,12,4) -180.0002 -DE/DX = 0.0 !
! D16 D(8,3,12,9) -0.0004 -DE/DX = 0.0 !
! D17 D(10,4,5,1) 180.0007 -DE/DX = 0.0 !
! D18 D(10,4,5,11) 0.0013 -DE/DX = 0.0 !
! D19 D(12,4,5,1) -0.0018 -DE/DX = 0.0 !
! D20 D(12,4,5,11) -180.0011 -DE/DX = 0.0 !
! D21 D(5,4,12,3) 0.0005 -DE/DX = 0.0 !
! D22 D(5,4,12,9) 180.0007 -DE/DX = 0.0 !
! D23 D(10,4,12,3) -180.0017 -DE/DX = 0.0 !
! D24 D(10,4,12,9) -0.0015 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
4. Calculation Summary
5. File Link
To access to file of optimization, click here.
Frequency Analysis of Pyridinium Molecule
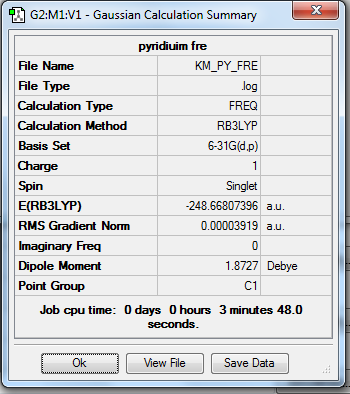
1. Calculation Summary
2. Real Output of Frequency Analysis of Pyridinium Molecule
Low frequencies --- -7.2125 0.0003 0.0005 0.0007 17.3350 18.5324
Low frequencies --- 392.4554 404.0615 620.4713
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A A
Frequencies -- 392.4553 404.0614 620.4713
Red. masses -- 2.9474 2.7453 6.2544
Frc consts -- 0.2675 0.2641 1.4187
IR Inten -- 0.9650 0.0000 0.0144
Atom AN X Y Z X Y Z X Y Z
1 6 0.00 0.00 0.25 0.00 0.00 0.00 0.39 0.00 0.00
2 6 0.00 0.00 -0.14 0.00 0.00 0.20 0.03 -0.23 0.00
3 6 0.00 0.00 -0.11 0.00 0.00 -0.19 -0.03 -0.20 0.00
4 6 0.00 0.00 -0.11 0.00 0.00 0.19 -0.03 0.20 0.00
5 6 0.00 0.00 -0.14 0.00 0.00 -0.20 0.03 0.23 0.00
6 1 0.00 0.00 0.61 0.00 0.00 0.00 0.39 0.00 0.00
7 1 0.00 0.00 -0.29 0.00 0.00 0.40 -0.25 -0.08 0.00
8 1 0.00 0.00 -0.20 0.00 0.00 -0.51 0.25 -0.01 0.00
9 1 0.00 0.00 0.46 0.00 0.00 0.00 -0.35 0.00 0.00
10 1 0.00 0.00 -0.20 0.00 0.00 0.51 0.25 0.01 0.00
11 1 0.00 0.00 -0.29 0.00 0.00 -0.40 -0.25 0.08 0.00
12 7 0.00 0.00 0.21 0.00 0.00 0.00 -0.34 0.00 0.00
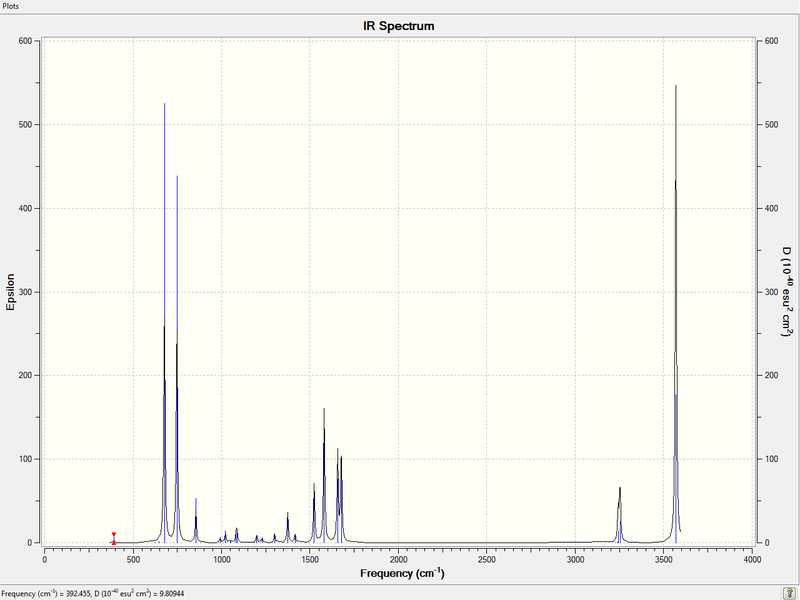
3. IR Spectrum of Pyridinium Molecule
4. File Link
To access the file link, click here
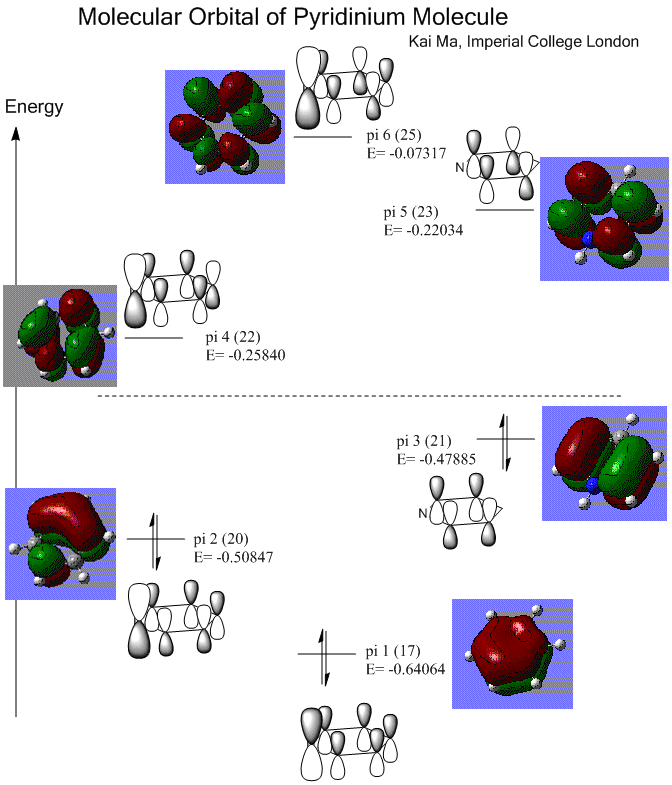
Molecular Orbital of Pyridinium Molecule
More electron density is pulled toward N atom. The molecule is still symmetric, but experienced a slight loss of symmetry comparing to benzene.
Isovalue = 0.03.
NBO analysis of Pyridinium Molecule
1. Calculation Summary
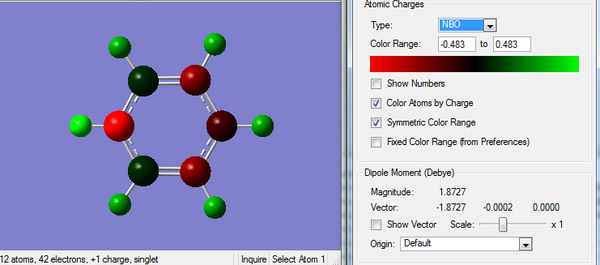
2. Charge Distribution of Pyridinium Molecule
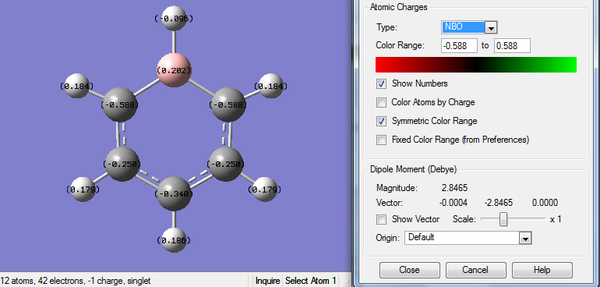
The analysis is carried out from previously optimized molecule with full population and full NBOs. The color range is set from -0.483 to +0.483 as shown below.
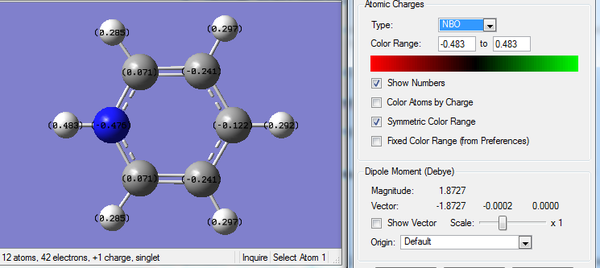
3. Charge Number of Pyridinium Molecule
4. Real Output of MO Analysis of Pyridinium Molecule
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
C 1 -0.12241 1.99913 4.10941 0.01386 6.12241
C 2 -0.24103 1.99912 4.22860 0.01331 6.24103
C 3 0.07098 1.99918 3.91068 0.01916 5.92902
C 4 0.07101 1.99918 3.91066 0.01916 5.92899
C 5 -0.24104 1.99912 4.22860 0.01331 6.24104
H 6 0.29170 0.00000 0.70718 0.00113 0.70830
H 7 0.29718 0.00000 0.70179 0.00103 0.70282
H 8 0.28493 0.00000 0.71397 0.00110 0.71507
H 9 0.48279 0.00000 0.51475 0.00246 0.51721
H 10 0.28493 0.00000 0.71397 0.00110 0.71507
H 11 0.29718 0.00000 0.70179 0.00103 0.70282
N 12 -0.47622 1.99937 5.46756 0.00929 7.47622
=======================================================================
* Total * 1.00000 11.99510 29.90895 0.09595 42.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98249) BD ( 1) C 1 - C 2
( 49.74%) 0.7053* C 1 s( 34.45%)p 1.90( 65.51%)d 0.00( 0.04%)
0.0000 0.5869 -0.0086 0.0005 -0.3935
0.0234 0.7063 0.0290 0.0000 0.0000
-0.0169 0.0000 0.0000 -0.0060 -0.0113
( 50.26%) 0.7089* C 2 s( 34.73%)p 1.88( 65.23%)d 0.00( 0.04%)
0.0000 0.5893 -0.0066 0.0009 0.4185
0.0371 -0.6897 0.0068 0.0000 0.0000
-0.0122 0.0000 0.0000 -0.0118 -0.0115
2. (1.98249) BD ( 1) C 1 - C 5
( 49.74%) 0.7053* C 1 s( 34.45%)p 1.90( 65.51%)d 0.00( 0.04%)
0.0000 0.5869 -0.0086 0.0005 -0.3934
0.0234 -0.7063 -0.0290 0.0000 0.0000
0.0169 0.0000 0.0000 -0.0060 -0.0113
( 50.26%) 0.7089* C 5 s( 34.73%)p 1.88( 65.23%)d 0.00( 0.04%)
0.0000 0.5893 -0.0066 0.0009 0.4184
0.0371 0.6898 -0.0068 0.0000 0.0000
0.0122 0.0000 0.0000 -0.0118 -0.0115
3. (1.54879) BD ( 2) C 1 - C 5
( 45.73%) 0.6762* C 1 s( 0.00%)p 1.00( 99.93%)d 0.00( 0.07%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9997 -0.0036
0.0000 -0.0241 -0.0101 0.0000 0.0000
( 54.27%) 0.7367* C 5 s( 0.00%)p 1.00( 99.94%)d 0.00( 0.06%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9997 -0.0080
0.0000 -0.0086 0.0228 0.0000 0.0000
4. (1.98141) BD ( 1) C 1 - H 6
( 64.64%) 0.8040* C 1 s( 31.07%)p 2.22( 68.89%)d 0.00( 0.03%)
-0.0003 0.5573 0.0131 -0.0007 0.8298
-0.0198 0.0001 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0153 -0.0101
( 35.36%) 0.5947* H 6 s( 99.94%)p 0.00( 0.06%)
0.9997 0.0018 -0.0242 0.0000 0.0000
5. (1.98297) BD ( 1) C 2 - C 3
( 49.58%) 0.7042* C 2 s( 33.47%)p 1.99( 66.48%)d 0.00( 0.05%)
0.0000 0.5784 -0.0119 -0.0002 -0.8145
-0.0194 -0.0009 0.0320 0.0000 0.0000
0.0047 0.0000 0.0000 0.0179 -0.0119
( 50.42%) 0.7100* C 3 s( 38.49%)p 1.60( 61.47%)d 0.00( 0.04%)
-0.0001 0.6204 -0.0023 0.0030 0.7832
0.0046 0.0142 0.0331 0.0000 0.0000
-0.0053 0.0000 0.0000 0.0168 -0.0095
6. (1.61445) BD ( 2) C 2 - C 3
( 52.23%) 0.7227* C 2 s( 0.00%)p 1.00( 99.94%)d 0.00( 0.06%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9997 -0.0068
0.0000 -0.0191 -0.0159 0.0000 0.0000
( 47.77%) 0.6912* C 3 s( 0.00%)p 1.00( 99.94%)d 0.00( 0.06%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9995 -0.0175
0.0000 0.0196 -0.0153 0.0000 0.0000
7. (1.97822) BD ( 1) C 2 - H 7
( 64.83%) 0.8052* C 2 s( 31.78%)p 2.15( 68.19%)d 0.00( 0.03%)
-0.0003 0.5636 0.0138 -0.0005 0.3988
-0.0072 0.7229 -0.0181 0.0000 0.0000
0.0109 0.0000 0.0000 -0.0085 -0.0099
( 35.17%) 0.5930* H 7 s( 99.94%)p 0.00( 0.06%)
0.9997 0.0016 -0.0116 -0.0208 0.0000
8. (1.98154) BD ( 1) C 3 - H 8
( 64.26%) 0.8016* C 3 s( 33.44%)p 1.99( 66.52%)d 0.00( 0.04%)
-0.0004 0.5780 0.0180 -0.0017 -0.4694
0.0193 0.6664 -0.0183 0.0000 0.0000
-0.0164 0.0000 0.0000 -0.0019 -0.0092
( 35.74%) 0.5978* H 8 s( 99.94%)p 0.00( 0.06%)
0.9997 0.0018 0.0128 -0.0209 0.0000
9. (1.98861) BD ( 1) C 3 - N 12
( 36.68%) 0.6057* C 3 s( 28.13%)p 2.55( 71.74%)d 0.00( 0.13%)
-0.0001 0.5293 -0.0335 -0.0013 -0.4042
-0.0563 -0.7417 -0.0277 0.0000 0.0000
0.0252 0.0000 0.0000 -0.0185 -0.0179
( 63.32%) 0.7957* N 12 s( 36.56%)p 1.73( 63.41%)d 0.00( 0.03%)
-0.0001 0.6046 -0.0037 0.0006 0.3658
-0.0187 0.7069 0.0132 0.0000 0.0000
0.0107 0.0000 0.0000 -0.0059 -0.0115
10. (1.98297) BD ( 1) C 4 - C 5
( 50.42%) 0.7100* C 4 s( 38.49%)p 1.60( 61.47%)d 0.00( 0.04%)
-0.0001 0.6204 -0.0023 0.0030 0.7832
0.0046 -0.0140 -0.0331 0.0000 0.0000
0.0053 0.0000 0.0000 0.0168 -0.0095
( 49.58%) 0.7042* C 5 s( 33.47%)p 1.99( 66.48%)d 0.00( 0.05%)
0.0000 0.5784 -0.0119 -0.0002 -0.8145
-0.0194 0.0007 -0.0320 0.0000 0.0000
-0.0047 0.0000 0.0000 0.0179 -0.0119
11. (1.98154) BD ( 1) C 4 - H 10
( 64.26%) 0.8016* C 4 s( 33.44%)p 1.99( 66.52%)d 0.00( 0.04%)
0.0004 -0.5780 -0.0180 0.0017 0.4693
-0.0193 0.6666 -0.0183 0.0000 0.0000
-0.0164 0.0000 0.0000 0.0019 0.0092
( 35.74%) 0.5978* H 10 s( 99.94%)p 0.00( 0.06%)
-0.9997 -0.0018 -0.0128 -0.0209 0.0000
12. (1.98861) BD ( 1) C 4 - N 12
( 36.68%) 0.6057* C 4 s( 28.13%)p 2.55( 71.74%)d 0.00( 0.13%)
-0.0001 0.5293 -0.0335 -0.0013 -0.4043
-0.0563 0.7416 0.0277 0.0000 0.0000
-0.0252 0.0000 0.0000 -0.0185 -0.0179
( 63.32%) 0.7957* N 12 s( 36.56%)p 1.73( 63.41%)d 0.00( 0.03%)
-0.0001 0.6047 -0.0037 0.0006 0.3660
-0.0187 -0.7069 -0.0132 0.0000 0.0000
-0.0107 0.0000 0.0000 -0.0059 -0.0115
13. (1.82447) BD ( 2) C 4 - N 12
( 28.55%) 0.5343* C 4 s( 0.00%)p 1.00( 99.83%)d 0.00( 0.17%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9991 0.0132
0.0000 -0.0102 0.0394 0.0000 0.0000
( 71.45%) 0.8453* N 12 s( 0.00%)p 1.00( 99.98%)d 0.00( 0.02%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9999 0.0036
0.0000 0.0128 -0.0077 0.0000 0.0000
14. (1.97822) BD ( 1) C 5 - H 11
( 64.83%) 0.8052* C 5 s( 31.78%)p 2.15( 68.19%)d 0.00( 0.03%)
0.0003 -0.5636 -0.0138 0.0005 -0.3989
0.0072 0.7228 -0.0181 0.0000 0.0000
0.0109 0.0000 0.0000 0.0085 0.0099
( 35.17%) 0.5930* H 11 s( 99.94%)p 0.00( 0.06%)
-0.9997 -0.0016 0.0116 -0.0208 0.0000
15. (1.98630) BD ( 1) H 9 - N 12
( 25.41%) 0.5041* H 9 s( 99.88%)p 0.00( 0.12%)
-0.9994 0.0064 -0.0342 0.0000 0.0000
( 74.59%) 0.8637* N 12 s( 26.82%)p 2.73( 73.16%)d 0.00( 0.02%)
0.0002 -0.5178 -0.0066 0.0013 0.8553
-0.0091 0.0001 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 -0.0115 0.0106
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (C5H6N)
1. BD ( 1) C 1 - C 2 1.98249 -0.90377 113(v),119(v),110(g),43(v)
107(g),112(g),63(v),62(v)
109(g)
2. BD ( 1) C 1 - C 5 1.98249 -0.90379 116(v),112(v),115(g),54(v)
106(g),119(g),33(v),32(v)
109(g)
3. BD ( 2) C 1 - C 5 1.54879 -0.44892 118(v),111(v),56(v),36(v)
4. BD ( 1) C 1 - H 6 1.98141 -0.71806 115(v),110(v),62(v),32(v)
107(g),106(g)
5. BD ( 1) C 2 - C 3 1.98297 -0.92650 120(v),109(v),106(g),23(v)
113(g),97(v),112(g),22(v)
114(g),96(v)
6. BD ( 2) C 2 - C 3 1.61445 -0.46666 108(v),118(v),99(v),111(g)
27(v)
7. BD ( 1) C 2 - H 7 1.97822 -0.71855 114(v),107(v),22(v),42(v)
106(g),110(g)
8. BD ( 1) C 3 - H 8 1.98154 -0.75115 117(v),106(v),32(v),110(g)
96(v)
9. BD ( 1) C 3 - N 12 1.98861 -1.06559 117(g),52(v),112(v),33(v)
116(v),54(v),110(g),120(g)
10. BD ( 1) C 4 - C 5 1.98297 -0.92648 120(v),109(v),107(g),23(v)
116(g),97(v),119(g),22(v)
117(g),96(v)
11. BD ( 1) C 4 - H 10 1.98154 -0.75115 114(v),107(v),62(v),115(g)
96(v)
12. BD ( 1) C 4 - N 12 1.98861 -1.06563 114(g),42(v),119(v),63(v)
113(v),43(v),115(g),120(g)
13. BD ( 2) C 4 - N 12 1.82447 -0.56812 111(v),108(v),44(v),66(v)
14. BD ( 1) C 5 - H 11 1.97822 -0.71855 117(v),106(v),22(v),52(v)
107(g),115(g)
15. BD ( 1) H 9 - N 12 1.98630 -0.89230 115(v),110(v),52(v),42(v)
16. CR ( 1) C 1 1.99914 -10.27397 63(v),33(v),110(v),115(v)
119(v),112(v),72(v)
17. CR ( 1) C 2 1.99913 -10.26482 23(v),43(v),42(v),113(v)
107(v),76(v),109(v),114(v)
18. CR ( 1) C 3 1.99918 -10.32332 33(v),117(v),120(v),106(v)
110(g),97(v),80(v),112(v)
19. CR ( 1) C 4 1.99918 -10.32333 63(v),114(v),120(v),107(v)
115(g),97(v),88(v),119(v)
20. CR ( 1) C 5 1.99913 -10.26482 23(v),54(v),52(v),116(v)
106(v),92(v),109(v),117(v)
21. CR ( 1) N 12 1.99937 -14.46219 54(v),43(v)
5. File Link
To access to the D-space link, click |here.
Analysis of Borazine Molecule
Optimization of Borazine Molecule
1. Optimized Borazine Molecule using 6-31G(d,p) Basis Set
test molecule |
2. Geometry Data
B-N Bond Length = 1.43 Å
N-H Bond Length = 1.01 Å
B-H Bond Length = 1.19 Å
Bond Angle = 119o and 121o
3. Real Output of Optimized Borazine Molecule
Item Value Threshold Converged?
Maximum Force 0.000093 0.000450 YES
RMS Force 0.000033 0.000300 YES
Maximum Displacement 0.000341 0.001800 YES
RMS Displacement 0.000096 0.001200 YES
Predicted change in Energy=-1.028617D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,7) 1.1949 -DE/DX = 0.0001 !
! R2 R(2,12) 1.0097 -DE/DX = 0.0 !
! R3 R(3,9) 1.1949 -DE/DX = 0.0001 !
! R4 R(4,11) 1.0097 -DE/DX = 0.0 !
! R5 R(5,8) 1.1949 -DE/DX = 0.0001 !
! R6 R(6,10) 1.0097 -DE/DX = 0.0 !
! R7 R(7,10) 1.4306 -DE/DX = 0.0 !
! R8 R(7,12) 1.4307 -DE/DX = 0.0 !
! R9 R(8,10) 1.4307 -DE/DX = -0.0001 !
! R10 R(8,11) 1.4306 -DE/DX = -0.0001 !
! R11 R(9,11) 1.4307 -DE/DX = -0.0001 !
! R12 R(9,12) 1.4307 -DE/DX = 0.0 !
! A1 A(1,7,10) 121.4414 -DE/DX = 0.0 !
! A2 A(1,7,12) 121.4399 -DE/DX = 0.0 !
! A3 A(10,7,12) 117.1187 -DE/DX = 0.0 !
! A4 A(5,8,10) 121.4373 -DE/DX = 0.0 !
! A5 A(5,8,11) 121.4436 -DE/DX = 0.0 !
! A6 A(10,8,11) 117.119 -DE/DX = 0.0001 !
! A7 A(3,9,11) 121.4337 -DE/DX = 0.0 !
! A8 A(3,9,12) 121.4348 -DE/DX = 0.0 !
! A9 A(11,9,12) 117.1314 -DE/DX = 0.0 !
! A10 A(6,10,7) 118.5584 -DE/DX = 0.0 !
! A11 A(6,10,8) 118.5563 -DE/DX = 0.0 !
! A12 A(7,10,8) 122.8853 -DE/DX = 0.0 !
! A13 A(4,11,8) 118.567 -DE/DX = 0.0 !
! A14 A(4,11,9) 118.5613 -DE/DX = 0.0 !
! A15 A(8,11,9) 122.8717 -DE/DX = 0.0 !
! A16 A(2,12,7) 118.5622 -DE/DX = 0.0 !
! A17 A(2,12,9) 118.5639 -DE/DX = 0.0 !
! A18 A(7,12,9) 122.8739 -DE/DX = 0.0 !
! D1 D(1,7,10,6) -0.0025 -DE/DX = 0.0 !
! D2 D(1,7,10,8) -179.9989 -DE/DX = 0.0 !
! D3 D(12,7,10,6) 179.9973 -DE/DX = 0.0 !
! D4 D(12,7,10,8) 0.0009 -DE/DX = 0.0 !
! D5 D(1,7,12,2) -0.0061 -DE/DX = 0.0 !
! D6 D(1,7,12,9) -179.9955 -DE/DX = 0.0 !
! D7 D(10,7,12,2) 179.9942 -DE/DX = 0.0 !
! D8 D(10,7,12,9) 0.0048 -DE/DX = 0.0 !
! D9 D(5,8,10,6) 0.0003 -DE/DX = 0.0 !
! D10 D(5,8,10,7) 179.9967 -DE/DX = 0.0 !
! D11 D(11,8,10,6) -179.9996 -DE/DX = 0.0 !
! D12 D(11,8,10,7) -0.0033 -DE/DX = 0.0 !
! D13 D(5,8,11,4) 0.0043 -DE/DX = 0.0 !
! D14 D(5,8,11,9) -179.9997 -DE/DX = 0.0 !
! D15 D(10,8,11,4) -179.9957 -DE/DX = 0.0 !
! D16 D(10,8,11,9) 0.0002 -DE/DX = 0.0 !
! D17 D(3,9,11,4) 0.0005 -DE/DX = 0.0 !
! D18 D(3,9,11,8) -179.9954 -DE/DX = 0.0 !
! D19 D(12,9,11,4) -179.9991 -DE/DX = 0.0 !
! D20 D(12,9,11,8) 0.005 -DE/DX = 0.0 !
! D21 D(3,9,12,2) 0.0034 -DE/DX = 0.0 !
! D22 D(3,9,12,7) 179.9928 -DE/DX = 0.0 !
! D23 D(11,9,12,2) -179.997 -DE/DX = 0.0 !
! D24 D(11,9,12,7) -0.0075 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
4. Calculation Summary
5. File Link
To access to the file link, click here
Frequency Analysis of Borazine Molecule
1. Calculation Summary
2. Real Output of Frequency Analysis of Borazine Molecule
Low frequencies --- -17.2100 -10.7688 -6.5918 -0.0013 -0.0013 -0.0013
Low frequencies --- 288.8517 289.6708 404.1673
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A A
Frequencies -- 288.8514 289.6706 404.1669
Red. masses -- 2.9273 2.9246 1.9257
Frc consts -- 0.1439 0.1446 0.1853
IR Inten -- 0.0000 0.0000 23.5576
Atom AN X Y Z X Y Z X Y Z
1 1 0.00 0.00 0.68 0.00 0.00 -0.13 0.00 0.00 0.53
2 1 0.00 0.00 -0.18 0.00 0.00 -0.20 0.00 0.00 0.16
3 1 0.00 0.00 -0.23 0.00 0.00 0.66 0.00 0.00 0.53
4 1 0.00 0.00 0.26 0.00 0.00 -0.05 0.00 0.00 0.16
5 1 0.00 0.00 -0.46 0.00 0.00 -0.53 0.00 0.00 0.53
6 1 0.00 0.00 -0.09 0.00 0.00 0.25 0.00 0.00 0.16
7 5 0.00 0.00 0.22 0.00 0.00 -0.04 0.00 0.00 0.10
8 5 0.00 0.00 -0.15 0.00 0.00 -0.17 0.00 0.00 0.10
9 5 0.00 0.00 -0.07 0.00 0.00 0.21 0.00 0.00 0.10
10 7 0.00 0.00 -0.08 0.00 0.00 0.23 0.00 0.00 -0.13
11 7 0.00 0.00 0.24 0.00 0.00 -0.05 0.00 0.00 -0.13
12 7 0.00 0.00 -0.16 0.00 0.00 -0.18 0.00 0.00 -0.13
3. IR Spectrum of Borazine Molecule
4. File Link
To access to the file link, click here.
Molecular Orbital of Borazine Molecule
There is a large difference in electronegativity between B atom and N atom. Therefore, electron density has been pulled towards N atoms to a large extent. Orbital degeneration appears as benzene.
Isovalue = 0.04.
NBO analysis of Borazine Molecule
1. Calculation Summary
2. Charge Distribution of Borazine Molecule
The analysis is carried out from previously optimized molecule with full population and full NBOs. The color range is set from -1.102 to +1.102 as shown below.
3. Charge Number of Borazine Molecule
4. Real Output of MO Analysis of Borazine Molecule
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
H 1 -0.07654 0.00000 1.07585 0.00069 1.07654
H 2 0.43199 0.00000 0.56573 0.00228 0.56801
H 3 -0.07655 0.00000 1.07586 0.00069 1.07655
H 4 0.43198 0.00000 0.56574 0.00228 0.56802
H 5 -0.07654 0.00000 1.07585 0.00069 1.07654
H 6 0.43198 0.00000 0.56574 0.00228 0.56802
B 7 0.74695 1.99917 2.23868 0.01521 4.25305
B 8 0.74700 1.99917 2.23862 0.01520 4.25300
B 9 0.74696 1.99917 2.23866 0.01521 4.25304
N 10 -1.10241 1.99943 6.09821 0.00478 8.10241
N 11 -1.10241 1.99943 6.09820 0.00478 8.10241
N 12 -1.10240 1.99943 6.09819 0.00478 8.10240
=======================================================================
* Total * 0.00000 11.99579 29.93532 0.06889 42.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98670) BD ( 1) H 1 - B 7
( 54.03%) 0.7351* H 1 s( 99.96%)p 0.00( 0.04%)
0.9998 0.0002 -0.0176 0.0077 0.0000
( 45.97%) 0.6780* B 7 s( 37.48%)p 1.67( 62.46%)d 0.00( 0.07%)
-0.0006 0.6120 0.0129 -0.0016 0.7239
-0.0247 -0.3160 0.0108 0.0000 0.0000
-0.0173 0.0000 0.0000 0.0161 -0.0098
2. (1.98495) BD ( 1) H 2 - N 12
( 28.08%) 0.5299* H 2 s( 99.91%)p 0.00( 0.09%)
0.9996 -0.0010 -0.0238 -0.0175 0.0000
( 71.92%) 0.8481* N 12 s( 22.82%)p 3.38( 77.15%)d 0.00( 0.03%)
-0.0002 0.4776 -0.0114 0.0006 0.7067
0.0105 0.5215 0.0077 0.0000 0.0000
0.0116 0.0000 0.0000 0.0036 -0.0119
3. (1.98670) BD ( 1) H 3 - B 9
( 54.03%) 0.7351* H 3 s( 99.96%)p 0.00( 0.04%)
0.9998 0.0002 0.0021 -0.0191 0.0000
( 45.97%) 0.6780* B 9 s( 37.47%)p 1.67( 62.46%)d 0.00( 0.07%)
-0.0006 0.6120 0.0129 -0.0016 -0.0883
0.0030 0.7849 -0.0267 0.0000 0.0000
-0.0052 0.0000 0.0000 -0.0230 -0.0098
4. (1.98495) BD ( 1) H 4 - N 11
( 28.08%) 0.5299* H 4 s( 99.91%)p 0.00( 0.09%)
-0.9996 0.0010 -0.0271 0.0118 0.0000
( 71.92%) 0.8481* N 11 s( 22.82%)p 3.38( 77.15%)d 0.00( 0.03%)
0.0002 -0.4776 0.0114 -0.0006 0.8049
0.0120 -0.3514 -0.0052 0.0000 0.0000
0.0089 0.0000 0.0000 -0.0083 0.0119
5. (1.98670) BD ( 1) H 5 - B 8
( 54.03%) 0.7351* H 5 s( 99.96%)p 0.00( 0.04%)
0.9998 0.0002 0.0154 0.0114 0.0000
( 45.97%) 0.6780* B 8 s( 37.48%)p 1.67( 62.46%)d 0.00( 0.07%)
-0.0006 0.6120 0.0129 -0.0016 -0.6355
0.0216 -0.4690 0.0160 0.0000 0.0000
0.0226 0.0000 0.0000 0.0070 -0.0098
6. (1.98495) BD ( 1) H 6 - N 10
( 28.08%) 0.5299* H 6 s( 99.91%)p 0.00( 0.09%)
-0.9996 0.0010 0.0033 -0.0293 0.0000
( 71.92%) 0.8481* N 10 s( 22.82%)p 3.38( 77.15%)d 0.00( 0.03%)
0.0002 -0.4776 0.0114 -0.0006 -0.0982
-0.0015 0.8727 0.0130 0.0000 0.0000
0.0027 0.0000 0.0000 0.0119 0.0119
7. (1.98438) BD ( 1) B 7 - N 10
( 23.53%) 0.4851* B 7 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
-0.0003 0.5587 -0.0174 0.0032 -0.6775
-0.0151 -0.4719 -0.0558 0.0000 0.0000
0.0412 0.0000 0.0000 0.0187 -0.0206
( 76.47%) 0.8745* N 10 s( 38.55%)p 1.59( 61.43%)d 0.00( 0.01%)
0.0000 0.6209 0.0043 -0.0001 0.6648
0.0017 0.4148 -0.0158 0.0000 0.0000
0.0065 0.0000 0.0000 0.0030 -0.0085
8. (1.82093) BD ( 2) B 7 - N 10
( 11.79%) 0.3433* B 7 s( 0.00%)p 1.00( 99.62%)d 0.00( 0.38%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9976 -0.0315
0.0000 -0.0545 -0.0283 0.0000 0.0000
( 88.21%) 0.9392* N 10 s( 0.00%)p 1.00(100.00%)d 0.00( 0.00%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 1.0000 -0.0003
0.0000 0.0000 0.0046 0.0000 0.0000
9. (1.98438) BD ( 1) B 7 - N 12
( 23.53%) 0.4851* B 7 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
0.0003 -0.5587 0.0174 -0.0032 0.1146
-0.0307 -0.8177 -0.0490 0.0000 0.0000
0.0155 0.0000 0.0000 0.0425 0.0206
( 76.47%) 0.8745* N 12 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 -0.6209 -0.0043 0.0001 -0.1480
-0.0127 0.7696 -0.0095 0.0000 0.0000
0.0025 0.0000 0.0000 0.0068 0.0085
10. (1.98438) BD ( 1) B 8 - N 10
( 23.53%) 0.4851* B 8 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
0.0003 -0.5587 0.0174 -0.0032 -0.7654
-0.0272 0.3095 0.0510 0.0000 0.0000
0.0290 0.0000 0.0000 -0.0347 0.0206
( 76.47%) 0.8745* N 10 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 -0.6209 -0.0043 0.0001 0.7404
-0.0019 -0.2567 0.0158 0.0000 0.0000
0.0046 0.0000 0.0000 -0.0055 0.0085
11. (1.98438) BD ( 1) B 8 - N 11
( 23.53%) 0.4851* B 8 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
0.0003 -0.5587 0.0174 -0.0032 0.0701
0.0408 -0.8227 -0.0410 0.0000 0.0000
0.0044 0.0000 0.0000 0.0450 0.0206
( 76.47%) 0.8745* N 11 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 -0.6209 -0.0043 0.0001 -0.0268
0.0145 0.7832 -0.0065 0.0000 0.0000
0.0007 0.0000 0.0000 0.0072 0.0085
12. (1.82090) BD ( 2) B 8 - N 11
( 11.79%) 0.3433* B 8 s( 0.00%)p 1.00( 99.62%)d 0.00( 0.38%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9976 -0.0315
0.0000 0.0028 0.0613 0.0000 0.0000
( 88.21%) 0.9392* N 11 s( 0.00%)p 1.00(100.00%)d 0.00( 0.00%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 1.0000 -0.0003
0.0000 0.0040 -0.0023 0.0000 0.0000
13. (1.98438) BD ( 1) B 9 - N 11
( 23.53%) 0.4851* B 9 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
-0.0003 0.5587 -0.0174 0.0032 -0.6508
-0.0578 -0.5080 0.0020 0.0000 0.0000
0.0445 0.0000 0.0000 0.0078 -0.0206
( 76.47%) 0.8745* N 11 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 0.6209 0.0043 -0.0001 0.5926
-0.0146 0.5129 0.0063 0.0000 0.0000
0.0071 0.0000 0.0000 0.0012 -0.0085
14. (1.98438) BD ( 1) B 9 - N 12
( 23.53%) 0.4851* B 9 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
0.0003 -0.5587 0.0174 -0.0032 -0.7475
-0.0559 0.3507 -0.0148 0.0001 0.0000
0.0367 0.0000 0.0000 -0.0263 0.0206
( 76.47%) 0.8745* N 12 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 -0.6209 -0.0043 0.0001 0.6917
-0.0129 -0.3683 -0.0094 0.0000 0.0000
0.0059 0.0000 0.0000 -0.0042 0.0085
15. (1.82091) BD ( 2) B 9 - N 12
( 11.79%) 0.3433* B 9 s( 0.00%)p 1.00( 99.62%)d 0.00( 0.38%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 -0.0001 0.0000 0.9976 -0.0315
0.0000 0.0517 -0.0331 0.0000 0.0000
( 88.21%) 0.9392* N 12 s( 0.00%)p 1.00(100.00%)d 0.00( 0.00%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 1.0000 -0.0003
0.0000 -0.0040 -0.0023 0.0000 0.0000
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (H6B3N3)
1. BD ( 1) H 1 - B 7 1.98670 -0.40394 115(v),119(v),76(v),96(v)
2. BD ( 1) H 2 - N 12 1.98495 -0.61482 118(v),112(v),119(g),114(g)
66(v),46(v)
3. BD ( 1) H 3 - B 9 1.98670 -0.40392 114(v),116(v),86(v),96(v)
4. BD ( 1) H 4 - N 11 1.98495 -0.61483 119(v),115(v),116(g),118(g)
56(v),66(v)
5. BD ( 1) H 5 - B 8 1.98670 -0.40394 118(v),112(v),86(v),76(v)
6. BD ( 1) H 6 - N 10 1.98495 -0.61480 114(v),116(v),112(g),115(g)
56(v),46(v)
7. BD ( 1) B 7 - N 10 1.98438 -0.68873 115(g),107(v),111(g),110(v)
57(v),116(v)
8. BD ( 2) B 7 - N 10 1.82093 -0.27140 117(v),62(v),58(v),43(v)
113(g)
9. BD ( 1) B 7 - N 12 1.98438 -0.68871 119(g),111(v),107(g),108(v)
67(v),118(v)
10. BD ( 1) B 8 - N 10 1.98438 -0.68868 112(g),109(v),111(g),106(v)
47(v),114(v)
11. BD ( 1) B 8 - N 11 1.98438 -0.68872 118(g),111(v),109(g),108(v)
67(v),119(v)
12. BD ( 2) B 8 - N 11 1.82090 -0.27139 120(v),72(v),68(v),35(v)
117(g)
13. BD ( 1) B 9 - N 11 1.98438 -0.68869 116(g),107(v),109(g),110(v)
57(v),115(v)
14. BD ( 1) B 9 - N 12 1.98438 -0.68871 114(g),109(v),107(g),106(v)
47(v),112(v)
15. BD ( 2) B 9 - N 12 1.82091 -0.27139 113(v),52(v),48(v),27(v)
120(g)
16. CR ( 1) B 7 1.99917 -6.65245 115(v),119(v),107(v),111(v)
17. CR ( 1) B 8 1.99917 -6.65247 112(v),118(v),109(v),111(v)
18. CR ( 1) B 9 1.99917 -6.65246 114(v),116(v),107(v),109(v)
19. CR ( 1) N 10 1.99943 -14.13097 47(v),57(v),112(g),115(g)
20. CR ( 1) N 11 1.99943 -14.13096 57(v),67(v),116(g),118(g)
21. CR ( 1) N 12 1.99943 -14.13097 47(v),67(v),114(g),119(g)
5. File Link
To access to the file link, click here.
Result and Discussion
General Result
1. Validity of Result
All of the four molecules present very similar total energies (difference less than 0.000001 a.u.) after frequency analysis comparing to the results in optimization. This means all the frequency analysis has been carried out correctly.
Also, all of the frequencies obtained have positive values, which indicates that the optimization analysis has been carried out successfully.
2. Total Energy
Total energy of benzene = -232.25820551
Total energy of boratabenzene = -219.02052984
Total energy of pyridinium = -248.66807396
Total energy of borazine = -242.68458727
By comparing the total energy of each molecule, they can be arranged in the following order from most stable to least stable:
pyridinium > borazine > benzene > boratabenzene
The validity of the result will be discussion later in the 'molecular orbital' session.
3. Symmetry Group
| Molecule | Symmetry group according to optimization | Literature point group |
| Benzene | C1 | D6h |
| Boratabenzene | C1 | Cs |
| Pyridinium | C1 | Cs |
| Borazine | C1 | D3h |
According to the table, all the symmetry groups have been assumed incorrectly because in order to get an energy minimum, all the symmetries have been switches off in the method.
Therefore, it makes no sense investigating into point groups further in this experiment.
4. Dipole Moment
| Molecule | Dipole moment according to optimization/ Debye | Literature dipole moment/ Debye |
| Benzene | 0.0001 | 0 |
| Boratabenzene | 2.8465 | N/A |
| Pyridinium | 1.8727 | N/A |
| Borazine | 0.0002 | 0.67[7] |
Similarly, since the method does not focus on symmetry, large error is involved in the prediction of dipole moment as well. However, although quantitative analysis on dipole moment failed, qualitative interpretation can still be made.
Benzene has a highly symmetric structure, thus no dipole moment.
In cases of boratabenzene and pyridinium, one of the C is replaced with other atoms with different electronegativity and an obvious increase in dipole moment is expected.
Borazine, which is also symmetric but has alternative distribution of N and B atoms, has a small dipole moment due to the electronegativity difference.
5. Bond Lengths and Bond Angles
All the calculations give bond angles near 120o, which is very close to literature.
| Molecule | Bond length according to optimization/ Å | Literature Bond length/ Å |
| Benzene |
C-C Bond Length = 1.40 |
C-C Bond Length = 1.40[8] |
| Boratabenzene |
C-B Bond length = 1.51 C-C Bond length = 1.40 |
C-B Bond length = 1.45-1.48 C-C Bond length = 1.38-1.41[9] |
| Pyridinium |
C-C Bond Length = 1.40 C-N Bond Length = 1.35 |
C-C Bond Length = 1.37 C-N Bond Length = 1.37 [10] |
| Borazine |
B-N Bond Length = 1.43 |
B-N Bond Length = 1.436 [11] |
The optimized bond lengths for four molecules are all close to literature. This proves that the method used in optimization is accurate and the results are reasonable.
The addition of electronegative atom (in this case, N atom) shortens the bond length to a small extent. Addition of electronpositive atom, in contrast, increases the bond length but the affection is negligible.
6. Aromaticity
All the four molecules are aromatic.
In terms of Huckel's rule, they all have:
1) planar structure;
2) 6 pi electrons (4n+2);
3) closed ring structure.
In terms of molecular orbital, their similarities are:
1) highly delocalized pi electron along the ring (not evenly distributed for all cases);
2) MOs contain formally only 2 electrons each.
Molecular Orbitals
1. HOMO, LUMO and Lowest pi Orbital
In order to maximize the visual effect of how the electron cloud is distributed, isovalue is set to 0.004, rather than the default value 0.02.
| Molecule | Benzene | Boratabenzene | Pyridinium | Borazine |
| Lowest pi Orbital | 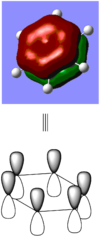 |
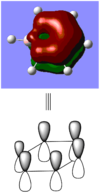 |
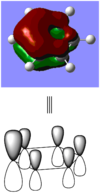 |
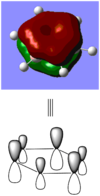
|
| HOMO | 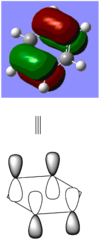 |
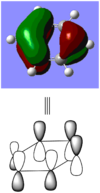 |
 |
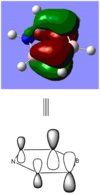
|
| LUMO | 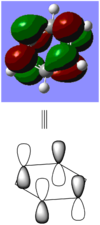 |
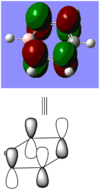 |
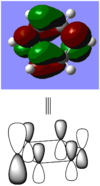 |
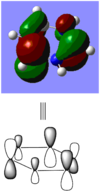
|
The lowest pi orbital, which is the totally bonding orbital, consist of 6 p orbitals from each ring atom perpendicular to the plane of ring in phase. They overlaps with each other and forms a delocalized system along the ring.
For benzene, all 6 p orbitals from carbons are identical and the electrons are distributed evenly. It provides large stabilization energy to benzene.
For boratabenzene, B atom is more electropositive than C atom, therefore has smaller p orbital. The electron density moved towards the rest part of the ring, i.e., the 5 C atoms. As from the diagram, there is an obvious weakening in electron density near B atom.
For pyridinium, conversely, N atom behaves more electronegative than C atom and pulls electron density towards itself.
For borazine, difference in electronegativity between B atom and N atom is large and results in uneven distribution of electron density. More electron is pulled towards N and forms large orbital. Though electrons are also delocalized in this molecule, the stabilization energy is weakened.
HOMO and LUMO in these molecules show very similar patterns of both bonding and anti-bonding interactions. Only slight difference is presented due to change of electronegativity in hetero-atoms. i.e., larger p orbital provided by electronegative N atom and smaller p orbital provided by electropositive B atom.
2. Energy level
| Molecule | Benzene | Boratabenzene | Pyridinium | Borazine |
| Energy level of Lowest pi Orbital | 17 | 17 | 17 | 17 |
| Energy level of HOMO | 20/ 21 (Degenerate) | 21 | 21 | 20/ 21 (Degenerate) |
| Energy level of LUMO | 22/ 23 (Degenerate) | 22 | 22 | 22/ 23 (Degenerate) |
| Energy level of Highest pi Orbital | 27 | 29 | 25 | 27 |
Energy levels of the lowest pi orbital, HOMO and LUMO in the four molecules are very similar despite the loss of degeneration in boratabenzene and pyridinium molecules. This is resulted from the loss of symmetry by replacing one of the C-H pattern by B-H or N-H pattern.
Energy level of the highest pi orbital, however, differs between boratabenzene and pyridinium due to the different sized p orbitals from B atom and N atom.
3. Charge Distribution and Charge Number
| Molecule | Benzene | Boratabenzene | Pyridinium | Borazine |
| Charge Number of C
(in order of most adjacent to hetero-atom to farthest away from hetero-atom) |
-0.239 | -0.588/ -0.258/ -0.340 | 0.071/ -0.241/ -0.122 | N/A |
| Charge Number of hetero-atom
(type of hetero-atom) |
N/A | 0.202 (B) | -0.476 (N) | 0.747 (B)/ -1.107 (N) |
| Charge Number of H connected to C
(in order of most adjacent to hetero-atom to farthest away from hetero-atom) |
0.239 | 0.184/ 0.179/ 0.189 | 0.285/ 0.297/ 0.292 | N/A |
| Charge Number of H connected to hetero-atom
(type of hetero-atom) |
N/A | -0.096 (B) | 0.483 (N) | -0.077 (B)/ 0.432 (N) |
| Sum of Charge Number | 0 | -1 | 1 | 0 |
The summary of charge numbers give a quantitatively way of looking at charge distribution. In this table, high symmetry of benzene is shown by the charge number of carbon being equal and exactly same in magnitude of hydrogen.
In boratabenzene, B atom gives a positive charge number, indicating its electropositive nature and lowering the charge number of C atoms and H atoms connected to C atoms. The extent to which charge number of C atom is lowered depends on the resonance forms which will be discussed in next session. Charge number of H atom connected to hetero-atom even increases from negative to a positive value in this molecule.
In pyridium, similar effect appears but in an opposite manner because N atom is more electronegative than C atom.
In borazine, the difference in electronegativity is enlarged. Symmetry is restored by the equality in charge number of H atoms connected to same hetero-atom.
4. Energy of Molecular Orbitals
| Molecule | Benzene | Boratabenzene | Pyridinium | Borazine |
| Energy of Lowest pi Orbital | -0.35998 | -0.13208 | -0.64064 | -0.36129 |
| Energy of HOMO | -0.24691 | 0.01094 | -0.47885 | -0.27590 |
| Energy of LUMO | 0.00267 | 0.21469 | -0.25840 | 0.02422 |
| Energy of Highest unoccupied pi Orbital | 0.16190 | 0.37027 | -0.07317 | 0.12496 |
A rapid rise in energy of MOs has been obtained when replacing one of C-H unit with the electropositive B-H unit. B atom has a natural negative charge but once the electron is delocalized, the electron density is pulled towards C atoms strongly, which makes the deficiency of electron in B atom even worse. Therefore, the molecule is overall destabilized.
In the opposite, N-H unit bring the energy of molecule down due to the electronegativity of N atom.
This result also proves the validity of the total energy calculation, that the order of stability is:
pyridinium > borazine > benzene > boratabenzene
Structure and Properties
Resonance forms of the four molecules have been compared and shown as below:
Despite confirming the relative stability between these molecules, it also tells the relationship with charge numbers as discussion previously. For example, in borazine, positive partial charge is distributed along N atoms while negative partial charge is distributed along B atoms. The results in a decrease in charge number of N atom and an increase in charge number of B atom. In boratabenzene and pyridinium molecules, the charge number of C atoms does not increase or decrease with the distance from hetero-atoms, but a 'jump' in the second C atom adjacent to hetero-atom. This is also presented in resonance forms as charges are usually distributed in those C atoms.
Conclusion
1. Aromaticity of benzene is not destroyed when replacing one C-H unit with another isoelectronic unit such as B-H or N-H.
2. When replaced by B-H or N-H, symmetry is lost due to electronegaticity difference. It disturbs the uniformly distributed electron, but not the delocalization nature.
3. Properties affected by replacing C-H unit include:
bond length: electronegative atom shortens the bond length. Bond angle is not affected.
charge distribution and charge number: electron is pulled towards more electronegative pattern, which has smaller charge number.
orbital degeneracy: due to loss of symmetry, degeneracy is lost.
LCAOs contributing to the MOs: more electropositive atom has smaller sized p orbital, thus smaller overall LCAO formed.
energy of the MOs: replacement of electronegative atom stabilizes the molecule while replacement of electropositive atom destabilizes the molecule.
ordering of MOs: highest unoccupied pi orbital orders differently in these molecules. The electronegative N atom raises the energy level while the electropositive B atom lowers the energy level.
4. In borazine, symmetry is partially restored by the recovering of orbital degenerate. However, charge distribution becomes even nonuniform due to large electronegativity difference between N and B.
References
- ↑ MAROULIS G. Bond Length Dependence of the Polarizability and Hyperpolarizability of Boron Hydride. International Journal of Quantum Chemistry. 2010.
- ↑ Julius Glaser GJ. On the Structures of the Hydrated Thallium(III) Ion and itsBromide Complexes in Aqueous Solution. Acta Chemica Scandinavica. 1982;A:125-135.
- ↑ John Morrison Galbraith GV, Henry F. Schaefer The vibrationalfrequencies of borane (BH3): A comparison of high level theoretical results. Journal of Molecular Structure. 1993;300:201-88.
- ↑ J. E. D. Davies et al., J. Chem. Soc., 1968, 2050
- ↑ I. J. Kurnig MMS, Steve Scheiner. Vibrational Frequencies and Intensities of H_Bonded Systems. 1:1 and 1:2 Complexes of NH3 and PH3 with HF. J Chem Phys. 1987;87:4.
- ↑ Harry.B.Gary. Chemical Bonds: An Introduction to Atomic and Molecular Structure.1994.
- ↑ H.J. Emelesus, A.G.Sharpe. Advances in Inorganic Chemistry and Radiochemistry,1963.
- ↑ March, J. “Advanced Organic Chemistry” 4th Ed. J. Wiley and Sons, 1992: New York. ISBN 0-471-60180-2.
- ↑ Diego A. Hoic, William M. Davis, and Gregory C. Fu. A Boron Analogue of Benzene: Synthesis, Structure, and Reactivity of 1-H-Boratabenzene. J Am Chem Soc. 1995;117:8480-1.
- ↑ Elschenbroich, C. Organometallchemie, 6th ed., p. 218, Vieweg+Teubner, 2008, ISBN 3-8351-0167-6
- ↑ Shriver and Atkins. Inorganic Chemistry (Fifth Edition). W. H. Freeman and Company, New York, 2010, pp 328.