Rep:Mod:jf301
Report
Optimisation of NH3
Calculation method: RB3LYP
Basis Set: 6-31G(d,p)
Final energy, E(RB3LYP): -56.55776873 a.u
RMS gradient: 0.00000485 a.u
Point group: C3V
NH bond length of final optimised structure: 1.01798 Å
HNH bond angle: 105.741 degrees
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986278D-10
Optimization completed.
-- Stationary point found.
Interactive NH3 model
Optimised NH molecule |
File:JFGRIFFIN NH3 OPTF POP.LOG
Vibrational analysis
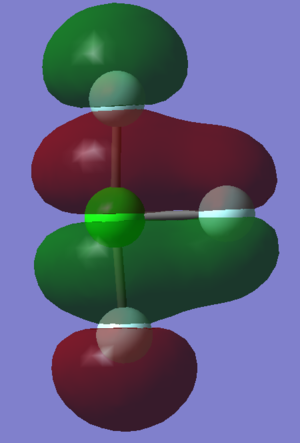
From the 3N-6 rule, (3*4)-6=6 vibrational modes are to be expected. There are two pairs of degenerate modes: modes 2 and 3 are degenerate, and modes 5 and 6 are degenerate. This can be determined from the frequency values in the table. Vibration modes 1, 2 and 3 are bending vibrations. Vibrations modes 4, 5 and 6 are stretching vibrations. Vibrational mode 4 is a highly symmetric stretch of all three NH bonds, simultaneously and in phase. Vibrational mode 1 is the "umbrella" mode; all three NH bonds are bending in the same direction. Two bands are to be expected in the IR spectrum of ammonia, due to the change in dipole moments in vibrational modes 1, 2 and 3 - modes 2 and 3 are degenerate and so contribute to the same signal in the spectrum.
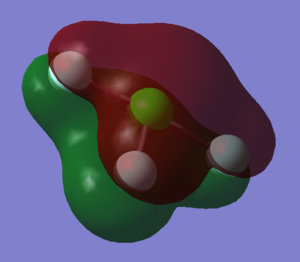
The charge associated with the Hydrogen atoms in the molecule is 0.375 and for the singular Nitrogen atom it is -1.125. Due to the symmetry of the molecule it is to be expected that all three Hydrogen atoms have the same charge, and that the sum of these positive charges must indeed be equal and opposite to the charge on Nitrogen, given that ammonia is a neutral molecule. It is also sensible that the program has predicted a negative charge for nitrogen, seeing as this is a more electronegative atom than hydrogen.
Optimisation of N2
Calculation method: RB3LYP
Basis Set: 6-31G(d,p)
Final energy, E(RB3LYP): -109.52412868 a.u
RMS gradient: 0.00000060 a.u
Point group: D*H
NN bond length of final optimised structure: 1.10550 Å
NN bond angle: Linear moecule, 180 degrees
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.400910D-13
Optimization completed.
-- Stationary point found.
Interactive N2 model
Optimised N molecule |
Vibrational Analysis
A vibration frequency of 2457.33 cm-1 is found for the molecule. This corresponds to the symmetric stretching of the bond. This vibration results in no change in dipole and as such will not be IR active.
Optimisation of H2
Calculation method: RB3LYP
Basis Set: 6-31G(d,p)
Final energy, E(RB3LYP): -1.17853936 a.u
RMS gradient: 0.00000017 a.u
Point group: D*H
NH bond length of final optimised structure: 0.74279 Å
HH bond angle: Linear molecule, 180 degrees
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
Interactive H2 model
Optimised H molecule |
Vibrational Analysis
A vibration frequency of 4465.68 cm-1 is found for the molecule. This corresponds to the symmetric stretching of the bond. This vibration results in no change in dipole and as such will not be IR active.
The Haber-Bosch Process
Energy Values in a.u
E(NH3)= -56.55776873 2*E(NH3)= -113.11553746 E(N2)= -109.52412868 E(H2)= -1.17853936 3*E(H2)= -3.53561808 ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.05579070 (8dp)
Energy Values in kJ/mol
E(NH3)= -148492.43 2*E(NH3)= -296984.87 E(N2)= -287555.62 E(H2)= -3094.26 3*E(H2)= -9282.77 ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -146.48 (2dp)
Energy calculation and analysis
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -146.48 (2dp)
This is the energy calculation for the formation of 2 moles of ammonia, therefore for the formation of 1 mole:
ΔE= -146.47848285/2 = -73.24 kJ/mol
Energy is lost as heat when moving from reactants to products; the Haber-Bosch process is an exothermic process. This indicates that the ammonia product is more stable than the gaseous reactants.
The literature value for the H2 bond strength is 435.7799 kJ/mol[1]. This differs from the computer approximated value of 3094.26 kJ/mol. This indicates that there is error in the approximation. A possible reason for this may be that in setting certain parameters for the approximation to be run at, the accuracy of the calculations may be limited. Additionally, literature values are usually reported at standard conditions but it is unknown how the computer program accounts for the effects of the atmosphere.
Optimisation of ClF3
Calculation method: RB3LYP
Basis Set: 6-31G(d,p)
Final energy, E(RB3LYP): -759.46531688 a.u
RMS gradient: 0.00002645 a.u
Point group: C2V
ClF bond lengths in final optimised structure: Two of length 1.72863 Å; these are the fluorine atoms bonded in the vertical axis. One of length 1.65146 Å, in the equatorial plane.
FClF bond angle: 87.140 degrees
FClF bond angle, using fluorine atoms in the vertical axis: 174.281 degrees
Item Value Threshold Converged?
Maximum Force 0.000050 0.000450 YES
RMS Force 0.000028 0.000300 YES
Maximum Displacement 0.000204 0.001800 YES
RMS Displacement 0.000134 0.001200 YES
Predicted change in Energy=-1.250248D-08
Optimization completed.
-- Stationary point found.
Interactive ClF3 model
Optimised ClF molecule |
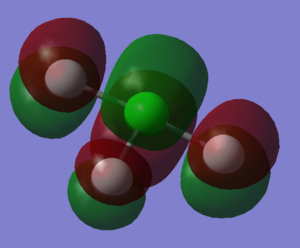
The molecule adopts a trigonal bipyramidal structure centered around the chlorine atom. Two fluorine atoms are bonded to opposite sides of the chlorine atom; this is the vertical axis. Another fluorine atom is bonded in the equatorial plane. Two lone pairs are also placed in orbitals in the equatorial plane. The molecule therefore adopts a 'T' shape.
The central chlorine atom has a calculated charge of 1.225. The two fluorine atoms bonded in the vertical axis are symmetrically identical and therefore have the same charge associated with them: -0.454. The fluorine atom bonded in the equatorial plane has a charge of -0.316. It is sensible for the vertical axis fluorine atoms to have a greater negative charge associated with them; this means electron density is more concentrated at 90 degrees away from the plane, and therefore from the electron lone pairs, rather than the ~120 degrees that the equatorial fluorine atom can provide.
Vibrational Analysis
By the 3N-6 rule for non-linear molecules, 3*(4)-6=6 vibrational modes are to be expected. Modes 1, 2 and 3 are bending modes and 4, 5 and 6 are stretching modes. The frequency values indicate that non of the modes are degenerate.