Rep:Mod:jem3v
Jenifer Mizen: Transition States and Reactivity
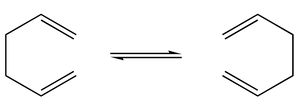
Cope Rearrangement

Firstly, 1,5-hexadiene with an approximately anitiperiplanar relationship between the four middle carbon atoms was optimised using HF/3-21G ( a Hartree-Fock method with the 3-21G basis set). The energy found by the summary was -231.68540au and the molecule was found to have C2h symmetry. The energy gradient and output files were both checked to ensure that convergence had been achieved.
Item Value Threshold Converged? Maximum Force 0.000058 0.000450 YES RMS Force 0.000023 0.000300 YES Maximum Displacement 0.001543 0.001800 YES RMS Displacement 0.000697 0.001200 YES Predicted change in Energy=-2.752863D-07 Optimization completed. -- Stationary point found.
This was repeated for a molecule in which the central carbons were gauche to each other. The energy was expected to be higher, as the a.p.p. conformation is generally the lowest in energy due to favourable interaction between the C-C (or C-H)σ-orbital and the neighbouring C-C (or C-H) σ*-orbital. The orbital overlap is best for the a.p.p. conformation compared to e.g. gauche.[2]The optimised energy was -231.69153 au and the symmetry was C2.
The two conformations are shown below:
A.p.p |
gauche |
The gauche structure corresponds to Gauche4 in Appendix 1. The initial antiperiplanar structure had the same symmetry as Anti3, but a different structure and slightly different energy. It was most similar to Anti1. A slightly different starting geometry was used and optimised to the anti1 conformation:
Antiperiplanar 1 |
This then gave the same point group and energy as anti1.
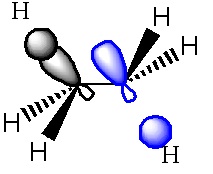
The Gauche structure was lower in energy than the antiperiplanar. This can be rationalised by considering the pi orbital interacting with the vicinal proton. Newman projections make it easier to visualise this interaction. Here, "D" corresponds to the minimum energy conformation (i.e. gauche 3 in the appendix 1).
.[3]
In the gauche form it was found that the distance between the terminal hydrogen (on the double bond) and a hydrogen on the third carbon atom was 2.44A, which corresponds to a van der Waals attractive interaction, (an A1,3 eclipsed conformation).
Overall, the σ-C-H/π*C=C interaction is responsible for the lower energy of the gauche form.

The lowest energy conformation was then optimised and is shown below:
Gauche 3 |
This has the same energy and point group as gauche 3 in appendix 1.
Next, the Ci anti2 conformation was optimised, and the symmetry checked to ensure it had stayed the same. The energy was found to be -231.69254au. This is the same as that given in the table. The molecule was then re-optimised, this time using the B3LYP/6-31G* level. The energy became more negative at -234.55970au.
| 1234 dihedral angle /o | 2345 dihedral angle /o | 1-2 & 5-6 bond length /A | 2-3 and 4-5 bond length /A | 3-4 bond length/A | |
|---|---|---|---|---|---|
| 1st optimisation | 114.7 | 180.0 | 1.32 | 1.51 | 1.55 |
| 2nd optimisation | 118.7 | 180.0 | 1.34 | 1.51 | 1.56 |
1st optimisation |
2nd optimisation |
For both, the 1234 dihedral angle was the same magnitude, but a different sign to the 3456 dihedral angle. This angle increased somewhat on the second optimisation, but there were no other particularly significant changes to the geometry.
Next, a frequency analysis was done using the same B3LYP/6-31G* level. No negative frequencies were found, with the lowest being at 71.69cm-1.
Sum of electronic and zero-point energies (potential energy at 0K) = -234.416244 au.
Sum of electronic and thermal energies (energy at 298.15K and 1atm, with contributions from translational, vibrational and rotational energy modes) = -234.408953 au.
Sum of electronic and thermal enthalpies (with correction for RT, H=E+RT) = -234.408009au.
Sum of electronic and thermal Free Energies (including entropy G=H-TS) = -234.447852.
PUT IR HERE RECALCULATE AT 0K IF TIME WHY AREN'T MY ANTI2 RESULTS THE SAME AS IN THE TABLE?
Optimising the Chair and Boat Transition Structures
Chair
Firstly, the allyl fragment was optimised (HF/3-21G). Then two of the optimised fragments were put together to give the approximate structure of the transition state. This was then optimised using two different methods:
1. Computing the force constant matrix (or the Hessian). This works well if the guess of the TS is very similar to the true structure.
HF/3-21G was used as before, and an Opt+Freq calculation done , with optimisation set to TS(Berny). To ensure that it did not crash if more than one imaginary frequency were found, "Opt=NoEigen" was added in the additional keywords section. An imaginary frequency was found at 818cm-1. The vibration is shown below:

TSforChairOptforwiki.mol |
On Gaussview, a dotted line is shown between the carbons, instead of one double and one single bond. This has not appeared using the jmol view, however the "single" and "double" bonds shown are actually the same length.
2. Freezing the reaction coordinate (so the rest of the molecule can be optimised), then unfreezing and re-optimising. This time, the distance between the terminal carbons of each fragment was set to 2.2A.
The molecule was then re-optimised to give a result identical to when using the previous method. The distance between the terminal allyl carbons i.e. where the new bond will form, was found to be 2.02A.
Boat
Next, the boat TS was optimised. The QST2 method was used. Two of the Ci anti2 molecules previously optimised were used and the atoms numbered so that the reactant and the product labelling matched. An Opt+Freq calculation was done, and the job failed, giving the following output:
Failed optimisation for boat TS.mol |
QST2 did not take into account the possibility of rotation about the central bonds, so failed to find the TS structure. The input was therefore modified so that the reactant (left hand side) and product (right hand side) had the following configurations:

The QST2 job was re-run. This gave error number 2070, and, when the .chk file was opened, Gaussview stopped responding. When the .out file was opened, it gave a "Gaussian error detected line number 1314" message, and the following structure:
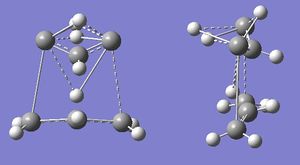
This has evidently not worked either. QST2 is very sensitive to how close the inputted structures are to the TS, so there may have been an error in the input file.
- ↑ Chemistry wiki Template:Www.ch.ic.ac.uk/wiki
- ↑ R. Pitzer, W. Lipscomb, J. Chem. Phys., 1963, 39, 1995. DOI:10.1063/1.1734572
- ↑ B. Gung, Z. Zhu, R. Fouch, J. am. Chem. Soc., 1995, 117, 1783-1788. DOI:10.1021/ja00111a016
- ↑ H. Rzepa,{{|http://vle.imperial.ac.uk/webct/cobaltMainFrame.dowebct}}
