Rep:Mod:jag310
Introduction
Computational chemistry can be very useful in allowing molecules that can't be characterised by experimental methods, to be analysed. It enables the location of transition states, and can provide important information about molecular orbitals, atomic interactions and dipole moments. In the following study, the program GaussView 5.0 was used to create and analyse molecules, using a variety of different basis sets and methodologies. In cases where more complex calculations were required, files were submitted to the HPC.
BH3 Optimisation
A BH3 molecule with trigonal planar geometry was drawn and the lengths of the B-H bonds were changed from the original 1.18A to 1.50A.
The H-B-H bond angle was 120.0 degrees.
Two different basis sets were used to optimise the borane molecule : 3-21G and 6-31G (d,p). Both were B3LYP
3-21G Basis Set
| BH3 | ||
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | B3YLP | |
| Basis Set | 3-21G | |
| Final Energy | -26.46 a.u. | |
| Gradient | 0.00020672 a.u. | |
| Dipole Moment | 0.00 Debye | |
| Point Group | D3h | |
| CPU Time | 9.0 seconds | |
| B-H Bond Length | 1.19 a.u. | |
| H-B-H Bond Angle | 120.0 | |
Item Value Threshold Converged? Maximum Force 0.000413 0.000450 YES RMS Force 0.000271 0.000300 YES Maximum Displacement 0.001610 0.001800 YES RMS Displacement 0.001054 0.001200 YES Predicted change in Energy=-1.071764D-06 Optimization completed. -- Stationary point found.
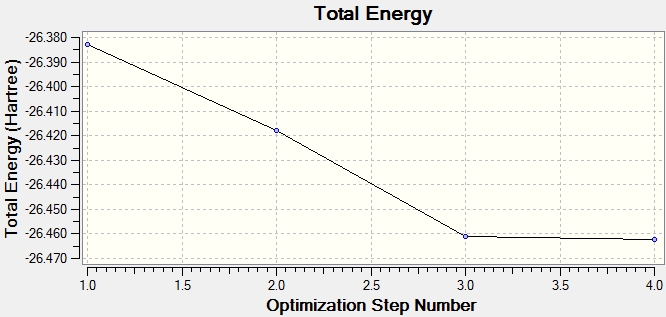
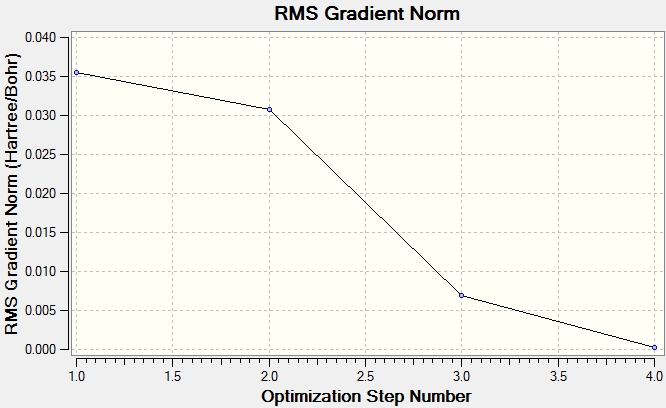
The optimisation of a molecule involves solving the Schrodinger equation for varying positions of the nuclei. The Total Energy graph is formed from Gaussview iteratively searching for the minimum energy structure, where the gradient (dE/dx) of the potential energy surface equals 0. The results from the calculation tabulate that the iterations have converged at dE/dx=0, showing that the molecule has been successfully optimised. The graph of the Root Mean Square (RMS) Gradient approaches zero as Gaussview gets closer to the energy minimum. The final structure has the lowest energy, corresponding to the smallest RMS gradient.
The above graphs and values of dE/dx won't be included for the rest of the calculations in this study.
6-31G Basis Set

| BH3 | ||
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | B3YLP | |
| Basis Set | 6-31G(d,p) | |
| Final Energy | -26.61 a.u. | |
| Gradient | 0.00000235 a.u. | |
| Dipole Moment | 0.00 Debye | |
| Point Group | D3h | |
| CPU Time | 10 seconds. | |
| B-H Bond Length | 1.19 a.u. | |
| H-B-H Bond Angle | 120.0 | |
Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.000019 0.001800 YES
RMS Displacement 0.000012 0.001200 YES
Predicted change in Energy=-1.304899D-10
Optimization completed.
-- Stationary point found.
TlBr3 Optimisation
TlBr3 was optimised with a medium level basis set. The high electron count of this molecule means that it displays relativistic effects. To account for this, a pseudo-potential has to be used, which implements the approximation that only valence electrons are involved in bonding interactions.
| TlBr3 | ||
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3YLP | |
| Basis Set | LANL2DZ | |
| Final Energy | -91.22 a.u. | |
| Gradient | 0.00275003 a.u. | |
| Dipole Moment | 0.00 Debye | |
| Point Group | D3h | |
| CPU Time | 29.5 seconds | |
| Tl-Br Bond Length | 2.65 a.u. | |
| Br-Tl-Br Bond Angle | 120.0 | |
The calculated bond length is fairly close to literature value of 2.52Å [1], which shows that the models used by the program are good approximations.
Item Value Threshold Converged? Maximum Force 0.000002 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000022 0.001800 YES RMS Displacement 0.000014 0.001200 YES Predicted change in Energy=-6.084017D-11 Optimization completed. -- Stationary point found.
BBr3 Optimisation
The calculation was published in D-space.
| BBr3 | ||
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3YLP | |
| Basis Set | GEN | |
| Final Energy | -64.43 a.u. | |
| Gradient | 0.01418127 a.u. | |
| Dipole Moment | 0.00 Debye | |
| Point Group | D3h | |
| CPU Time | 18.1 seconds | |
| B-Br Bond Length | 1.93 a.u. | |
| H-B-H Bond Angle | 120.0 | |
The calculated bond length is fairly close to literature value of 1.88Å [2], which shows that the models used by the program are good approximations.
Item Value Threshold Converged? Maximum Force 0.000008 0.000450 YES RMS Force 0.000005 0.000300 YES Maximum Displacement 0.000036 0.001800 YES RMS Displacement 0.000023 0.001200 YES Predicted change in Energy=-4.026911D-10 Optimization completed. -- Stationary point found.
Structure Comparison
| Atom | Electron Configuration |
|---|---|
| B | [He] 2s2 2p1 |
| Br | [Ar] 4s2 3d10 4p5 |
| H | 1s1 |
| Tl | [Xe] 4f14 5d10 6s2 6p1 |
| Molecule | Bond Distance (a.u.) |
|---|---|
| BH3 | 1.19 |
| BBr3 | 1.93 |
| TlBr3 | 2.65 |
Tl has larger, most diffuse valence orbitals than Boron, with a greater amount of shielding. This means that Tl gives poorer overlap with a substituent and so the longest bond distance is between Tl and Br. It is the same situation for Br in comparison to H. Br has more diffuse orbitals, with greater shielding than H, so the B-Br bond length is greater than that of B-H.
Both Br and H are non-metals with electronegativities of 2.96 and 2.20 respectively (Pauling Scale), compared to B with 2.04. The bonding character in BH3 will be covalent with comparatively little polarity in the B-H bonds, compared to BBr3 which is also covalently bonded but with a greater polarity in the B-Br bonds.
BH3 Frequency Analysis

| BH3 | ||
|---|---|---|
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3YLP | |
| Basis Set | 6-31G(d,p) | |
| Final Energy | -26.61 a.u. | |
| Gradient | 0.00000237 a.u. | |
| Dipole Moment | 0.00 Debye | |
| Point Group | D3h | |
| CPU Time | 12.0 seconds | |
| B-H Bond Length | 1.19 a.u. | |
| H-B-H Bond Angle | 120.0 | |
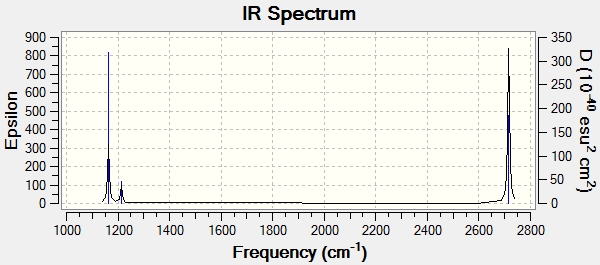
Low frequencies --- -0.9033 -0.7343 -0.0054 6.7375 12.2491 12.2824 Low frequencies --- 1163.0003 1213.1853 1213.1880
Item Value Threshold Converged? Maximum Force 0.000005 0.000450 YES RMS Force 0.000002 0.000300 YES Maximum Displacement 0.000019 0.001800 YES RMS Displacement 0.000009 0.001200 YES Predicted change in Energy=-1.323374D-10 Optimization completed. -- Stationary point found.
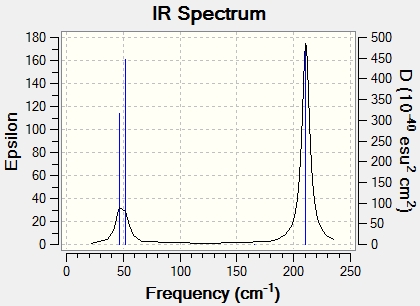
Main peaks (Frequency/cm-1): 1163, 1213, 2715
There are 6 vibrational modes, but the predicted IR spectrum only has 3 peaks. This is because modes 1 and 2 are degenerate, so they only produce one peak, because they have the same wavenumber. Mode 4 has no intensity (0.00) because it is a fully symmetrical stretch, resulting in no change of the dipole moment. Modes 5 and 6 are again degenerate, having the same wavenumber and so again only contribute 1 peak between them. This gives 3 peaks in total, as seen in the predicted spectrum.
TlBr3 Frequency
The calculation was published in D-space.

| TlBr3 | ||
|---|---|---|
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3YLP | |
| Basis Set | LANL2DZ | |
| Final Energy | -91.22 a.u. | |
| Gradient | 0.00000088 a.u. | |
| Dipole Moment | 0.00 Debye | |
| Point Group | D3h | |
| CPU Time | 15.7 seconds | |
| Tl-Br Bond Length | 2.65 a.u. | |
| Br-Tl-Br Bond Angle | 120.0 | |
AS with BH3, the 6 vibrational modes only equate to 3 peaks in the predicted IR spectrum, due to degeneracy and the lack of dipole moment change in mode 4.
Item Value Threshold Converged? Maximum Force 0.000002 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000022 0.001800 YES RMS Displacement 0.000011 0.001200 YES Predicted change in Energy=-5.660901D-11 Optimization completed. -- Stationary point found.
Low frequencies --- -3.4213 -0.0026 -0.0004 0.0015 3.9367 3.9367 Low frequencies --- 46.4289 46.4292 52.1449
Main peaks (Frequency/cm-1): 46, 52, 211
Comparison of TlBr3 and BH3 Vibrational Frequencies
| Vibrational Mode | BH3 Frequency (cm-1) | TlBr3 Frequency (cm-1) |
|---|---|---|
| 1 | 1163.00 | 46.43 |
| 2 | 1213.19 | 46.43 |
| 3 | 1213.19 | 52.14 |
| 4 | 2582.26 | 165.27 |
| 5 | 2715.43 | 210.69 |
| 6 | 2715.43 | 210.69 |
The vibrational frequencies can be approximated using Hooke's Law, which shows that the frequency is proportional to the square root of the bond constant and 1/square root of the reduced mass. This explains why the BH3 vibrational frequencies are all at higher wavenumbers than those of TlBr3; the more diffuse orbitals of the Tl and Br atoms gives a poorer orbital overlap, and so a smaller bond constant and the higher atomic weight of the Tl and Br atoms, gives them a higher reduced mass.
The same basis set and method has to be used for the optimisation and frequency analysis calculations due to the total energy being dependent on which basis set is used. For comparison, the same number of atoms is required, with the same basis-set throughout. Non-linear molecules have 3N-6 vibrational frequencies. The low frequencies represent the '-6' part of the vibrational frequencies. The positive values from this onwards, are the 'real' vibrational frequencies.
BH3 Population Analysis
The calculation was published in D-space.

| BH3 | ||
|---|---|---|
| File Type | .log | |
| Calculation Type | SP | |
| Calculation Method | RB3YLP | |
| Basis Set | 6-31G(d,p) | |
| Final Energy | -26.61 a.u. | |
| Gradient | 0.0000000 a.u. | |
| Dipole Moment | 0.00 Debye | |
| Point Group | D3h | |
| CPU Time | 10.5 seconds | |
| B-H Bond Length | 1.19 a.u. | |
| H-B-H Bond Angle | 120.0 | |
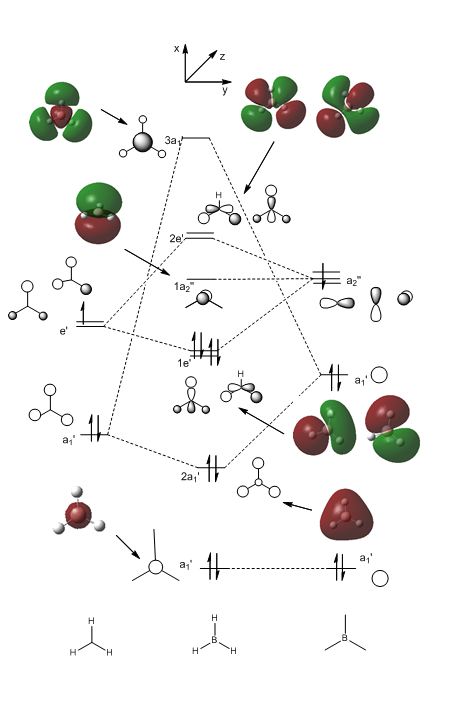
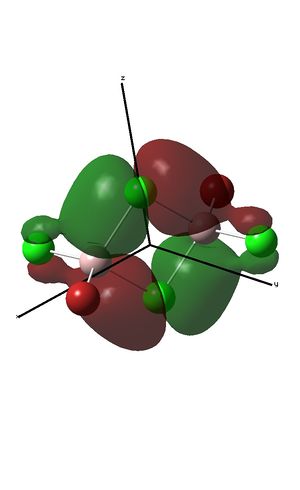
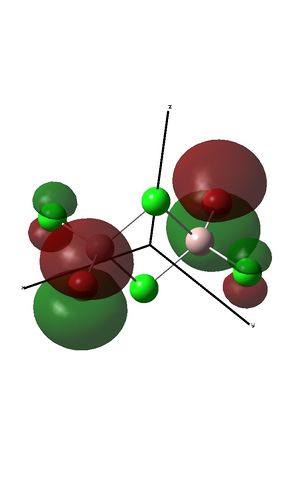
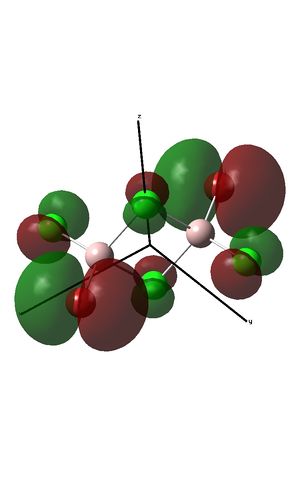
The fragment orbitals used to create the MO diagram were H3, with a central B atom. For the H3 fragment, the lowest energy fragment orbital is bonding and symmetric, which has a1' symmetry. There are two degenerate higher energy FOs of e' symmetry. For the B atom, the lowest energy orbital is the s orbital, which has a1' symmetry. The px and py orbitals are degenerate with e' symmetry and the pz orbital has a2" symmetry. B and H have similar electronegativities and so the two a1' symmetry orbitals combine with a large splitting energy. The e' orbitals also combine, but with with a smaller splitting energy. The a2" orbitals remain non-bonding.
NH3 NBO Analysis
NH3 Optimisation

| NH3 | ||
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3YLP | |
| Basis Set | 6-31G(d,p) | |
| Final Energy | -56.56 a.u. | |
| Gradient | 0.00000885 a.u. | |
| Dipole Moment | 1.85 Debye | |
| Point Group | C1 | |
| CPU Time | 15.0 seconds | |
| N-H Bond Length | 1.02 a.u. | |
| H-N-H Bond Angle | 105.7 | |
Item Value Threshold Converged? Maximum Force 0.000024 0.000450 YES RMS Force 0.000012 0.000300 YES Maximum Displacement 0.000079 0.001800 YES RMS Displacement 0.000053 0.001200 YES Predicted change in Energy=-1.629731D-09 Optimization completed. -- Stationary point found.
NH3 Frequency Analysis
| NH3 | ||
|---|---|---|
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3YLP | |
| Basis Set | 6-31G(d,p) | |
| Final Energy | -56.56 a.u. | |
| Gradient | 0.00000888 a.u. | |
| Dipole Moment | 1.85 Debye | |
| Point Group | C1 | |
| CPU Time | 15.0 seconds | |
| N-H Bond Length | 1.02 a.u. | |
| H-N-H Bond Angle | 105.7 | |
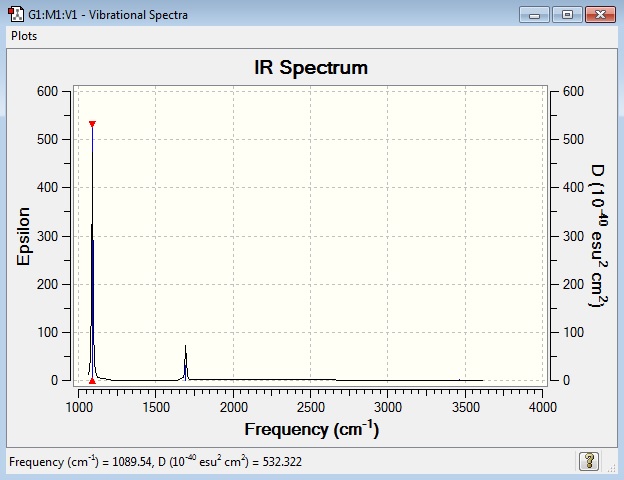
Low frequencies --- -30.7764 -0.0019 -0.0015 -0.0011 20.3142 28.2484 Low frequencies --- 1089.5557 1694.1237 1694.1868
Item Value Threshold Converged? Maximum Force 0.000021 0.000450 YES RMS Force 0.000009 0.000300 YES Maximum Displacement 0.000077 0.001800 YES RMS Displacement 0.000039 0.001200 YES Predicted change in Energy=-1.603126D-09 Optimization completed. -- Stationary point found.
Main peaks (Frequency/cm-1): 1090, 1694
NH3 Population Analysis
The calculation was published in D-space.
| NH3 | ||
|---|---|---|
| File Type | .log | |
| Calculation Type | SP | |
| Calculation Method | RB3YLP | |
| Basis Set | 6-31G(d,p) | |
| Final Energy | -56.56 a.u. | |
| Gradient | 0.00000888 a.u. | |
| Dipole Moment | 1.85 Debye | |
| Point Group | C1 | |
| CPU Time | 6.5 seconds | |
| N-H Bond Length | 1.02 a.u. | |
| H-N-H Bond Angle | 105.7 | |
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
N 1 -1.12515 1.99982 6.11104 0.01429 8.12515
H 2 0.37505 0.00000 0.62250 0.00246 0.62495
H 3 0.37505 0.00000 0.62250 0.00246 0.62495
H 4 0.37505 0.00000 0.62249 0.00246 0.62495
=======================================================================
* Total * 0.00000 1.99982 7.97852 0.02166 10.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.99909) BD ( 1) N 1 - H 2
( 68.83%) 0.8297* N 1 s( 24.87%)p 3.02( 75.05%)d 0.00( 0.09%)
-0.0001 -0.4986 -0.0059 0.0000 -0.2910
0.0052 0.8155 0.0277 0.0000 0.0000
0.0281 0.0000 0.0000 0.0032 0.0082
( 31.17%) 0.5583* H 2 s( 99.91%)p 0.00( 0.09%)
-0.9996 0.0000 0.0072 -0.0289 0.0000
2. (1.99909) BD ( 1) N 1 - H 3
( 68.83%) 0.8297* N 1 s( 24.86%)p 3.02( 75.05%)d 0.00( 0.09%)
0.0001 0.4986 0.0059 0.0000 0.2910
-0.0052 0.4077 0.0138 0.7062 0.0240
0.0140 0.0243 0.0076 0.0033 0.0031
( 31.17%) 0.5583* H 3 s( 99.91%)p 0.00( 0.09%)
0.9996 0.0000 -0.0072 -0.0145 -0.0250
3. (1.99909) BD ( 1) N 1 - H 4
( 68.83%) 0.8297* N 1 s( 24.87%)p 3.02( 75.05%)d 0.00( 0.09%)
0.0001 0.4986 0.0059 0.0000 0.2909
-0.0052 0.4077 0.0138 -0.7062 -0.0239
0.0140 -0.0243 -0.0076 0.0033 0.0031
( 31.17%) 0.5583* H 4 s( 99.91%)p 0.00( 0.09%)
0.9996 0.0000 -0.0072 -0.0145 0.0250
4. (1.99982) CR ( 1) N 1 s(100.00%)
1.0000 -0.0002 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
5. (1.99721) LP ( 1) N 1 s( 25.38%)p 2.94( 74.52%)d 0.00( 0.10%)
0.0001 0.5036 -0.0120 0.0000 -0.8618
0.0505 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 -0.0269 0.0155
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (H3N)
1. BD ( 1) N 1 - H 2 1.99909 -0.60417
2. BD ( 1) N 1 - H 3 1.99909 -0.60417
3. BD ( 1) N 1 - H 4 1.99909 -0.60416
4. CR ( 1) N 1 1.99982 -14.16768
5. LP ( 1) N 1 1.99721 -0.31756 24(v),16(v),20(v),17(v)
21(v),25(v)
Calculation of charge distribution between -1.000 to +1.000 was colour coordinated, with red indicating an area of negative charge region and green a positively charged region.
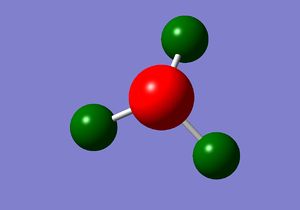
The relative charges were -1.125 for the central N atom and +0.375 for the H atoms.
NH3BH3
NH3BH3 Optimisation
3-21 G Basis Set
| NH3BH3 | ||
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3YLP | |
| Basis Set | 2-31G | |
| Final Energy | -82.77 a.u. | |
| Gradient | 0.00003006 a.u. | |
| Dipole Moment | 5.84 Debye | |
| Point Group | C1 | |
| CPU Time | 42.0 seconds. | |
| N-H Bond Length | 1.03 a.u. | |
| H-N-H Bond Angle | 109.4 | |
| B-H Bond Length | 1.21 a.u. | |
| H-B-H Bond Angle | 113.6 | |
Item Value Threshold Converged? Maximum Force 0.000094 0.000450 YES RMS Force 0.000030 0.000300 YES Maximum Displacement 0.000419 0.001800 YES RMS Displacement 0.000178 0.001200 YES Predicted change in Energy=-5.742850D-08 Optimization completed. -- Stationary point found.
6-31G Basis Set
| NH3BH3 | ||
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3YLP | |
| Basis Set | 6-31G(d,p) | |
| Final Energy | -83.22 a.u. | |
| Gradient | 0.00005649 a.u. | |
| Dipole Moment | 5.56 Debye | |
| Point Group | C1 | |
| CPU Time | 50.0 seconds. | |
| N-H Bond Length | 1.02 a.u. | |
| H-N-H Bond Angle | 107.9 | |
| B-H Bond Length | 1.21 a.u. | |
| H-B-H Bond Angle | 113.9 | |
Item Value Threshold Converged? Maximum Force 0.000132 0.000450 YES RMS Force 0.000037 0.000300 YES Maximum Displacement 0.001192 0.001800 YES RMS Displacement 0.000531 0.001200 YES Predicted change in Energy=-1.177617D-07 Optimization completed. -- Stationary point found.
Frequency Analysis
File:JULIUSGUTH NH3BH3FREQ.LOG
| NH3BH3 | ||
|---|---|---|
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3YLP | |
| Basis Set | 6-31G(d,p) | |
| Final Energy | -83.22 a.u. | |
| Gradient | 0.00005654 a.u. | |
| Dipole Moment | 5.56 Debye | |
| Point Group | C1 | |
| CPU Time | 69.0 seconds | |
| N-H Bond Length | 1.018 a.u. | |
| H-N-H Bond Angle | 107.9 | |
| B-H Bond Length | 1.21 a.u. | |
| H-B-H Bond Angle | 113.9 | |
Low frequencies --- -0.0012 -0.0010 -0.0009 3.5300 15.6870 20.3924 Low frequencies --- 263.3630 631.3094 638.1307
Item Value Threshold Converged? Maximum Force 0.000254 0.000450 YES RMS Force 0.000057 0.000300 YES Maximum Displacement 0.001412 0.001800 YES RMS Displacement 0.000694 0.001200 YES Predicted change in Energy=-2.141505D-07 Optimization completed. -- Stationary point found.
Main peaks (Frequency/cm-1): 638, 1069, 1204, 1329, 1676, 2472, 2533, 3581
Association Energy of NH3BH3
E(BH3) = -26.61 a.u.
E(NH3) = -56.56 a.u.
E(BH3NH3) = -83.22 a.u.
ΔE = E(NH3BH3)-[E(NH3)+E(BH3)]
ΔE = -83.22 a.u. - -83.17
ΔE = -0.05 a.u.
1 a.u. = 2625.5 kJ mol-1
Enthalpy of formation of ammonia-borane ΔHf = -0.05 a.u. * 2625.5 = -131.3 kJ mol-1
This value is lower than the literature value of -172.1 kJ mol-1
Lewis Acid Mini project
Cl2Al(μ-Br2)AlCl2 isomer
Optimisation and pseudo-potential
File:Al2br2cl4optimised631gpseudo.log

| Al2Br2Cl4 | ||
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | GEN | |
| Final Energy | -2352.40630798 a.u. | |
| Gradient | 0.00000396 a.u. | |
| Dipole Moment | 0.00 Debye | |
| Point Group | D2h | |
| CPU Time | 3 minutes 21.0 seconds | |
Item Value Threshold Converged?
Maximum Force 0.000007 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.000154 0.001800 YES
RMS Displacement 0.000058 0.001200 YES
Predicted change in Energy=-1.207258D-09
Optimization completed.
-- Stationary point found.
Frequency Analysis
Low frequencies --- -5.1810 -5.0302 -3.2289 -0.0003 0.0012 0.0031 Low frequencies --- 14.8284 63.2817 86.0839
Item Value Threshold Converged?
Maximum Force 0.000013 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000209 0.001800 YES
RMS Displacement 0.000099 0.001200 YES
Predicted change in Energy=-2.119810D-09
Optimization completed.
-- Stationary point found.
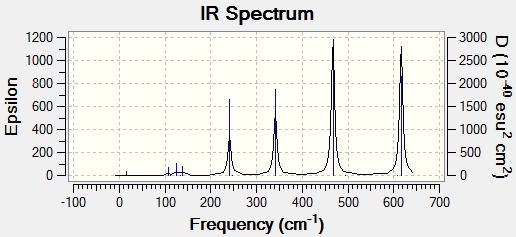
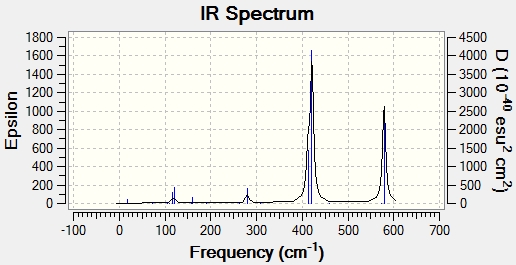
Main peaks (Frequency/cm-1): 241, 341, 467, 616
BrClAl(μ-Br,Cl)AlCl2
Optimisation and pseudo-potential
File:Al2br2cl4 2ndisomerpseudoout.log
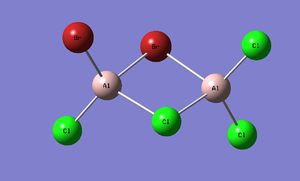
| Al2Br2Cl4 | ||
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | GEN | |
| Final Energy | -2352.41109944 a.u. | |
| Gradient | 0.00001557 a.u. | |
| Dipole Moment | 0.1386 Debye | |
| Point Group | D2h | |
| CPU Time | 4 minutes 16.8 seconds | |
Item Value Threshold Converged?
Maximum Force 0.000035 0.000450 YES
RMS Force 0.000014 0.000300 YES
Maximum Displacement 0.000509 0.001800 YES
RMS Displacement 0.000182 0.001200 YES
Predicted change in Energy=-2.015419D-08
Optimization completed.
-- Stationary point found.
Frequency Analysis
Low frequencies --- -2.2907 0.0011 0.0016 0.0019 1.2447 3.3231 Low frequencies --- 17.1610 55.9531 80.0563
Item Value Threshold Converged? Maximum Force 0.000034 0.000450 YES RMS Force 0.000016 0.000300 YES Maximum Displacement 0.001349 0.001800 YES RMS Displacement 0.000527 0.001200 YES Predicted change in Energy=-3.663910D-08 Optimization completed. -- Stationary point found.
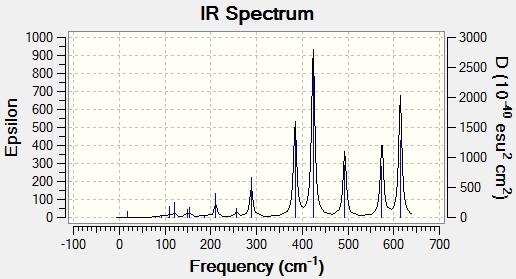
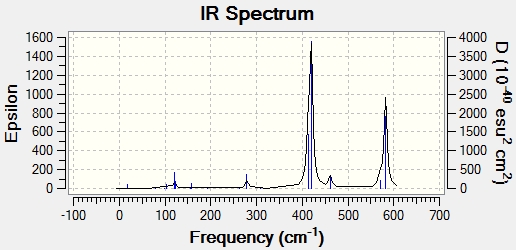
Main peaks (Frequency/cm-1): 289, 384, 424, 493, 574, 614
Trans-BrClAl(μ-Cl2)AlClBr
Optimisation and pseudo-potential
File:Al isomer3outputpseudo.log
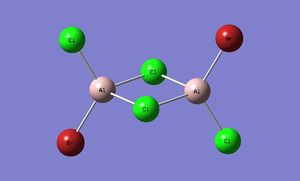
| Al2Br2Cl4 | ||
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | GEN | |
| Final Energy | -2352.41629858 a.u. | |
| Gradient | 0.00001567 a.u. | |
| Dipole Moment | 0.00 Debye | |
| Point Group | D2h | |
| CPU Time | 4 minutes 6.2 seconds | |
Item Value Threshold Converged?
Maximum Force 0.000039 0.000450 YES
RMS Force 0.000015 0.000300 YES
Maximum Displacement 0.000467 0.001800 YES
RMS Displacement 0.000168 0.001200 YES
Predicted change in Energy=-2.435287D-08
Optimization completed.
-- Stationary point found.
Frequency Analysis
Low frequencies --- -4.7965 0.0020 0.0024 0.0030 1.4551 2.2602 Low frequencies --- 18.1749 49.1211 73.0074
Item Value Threshold Converged?
Maximum Force 0.000050 0.000450 YES
RMS Force 0.000016 0.000300 YES
Maximum Displacement 0.000576 0.001800 YES
RMS Displacement 0.000258 0.001200 YES
Predicted change in Energy=-3.235482D-08
Optimization completed.
-- Stationary point found.
Main peaks (Frequency/cm-1): 421, 579
Cis-BrClAl(μ-Cl2)AlClBr
Optimisation and pseudo-potential
File:Al pseudooutputisomer4.log
| Al2Br2Cl4 | ||
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | GEN | |
| Final Energy | -2352.41626677 a.u. | |
| Gradient | 0.00001470 a.u. | |
| Dipole Moment | 0.1658 Debye | |
| Point Group | D2h | |
| CPU Time | 4 minutes 8.1 seconds. | |
Item Value Threshold Converged?
Maximum Force 0.000040 0.000450 YES
RMS Force 0.000016 0.000300 YES
Maximum Displacement 0.001359 0.001800 YES
RMS Displacement 0.000424 0.001200 YES
Predicted change in Energy=-2.577003D-08
Optimization completed.
-- Stationary point found.
Frequency Analysis
Low frequencies --- -3.8191 -2.2355 -0.0023 -0.0015 0.0007 1.3869 Low frequencies --- 17.2012 50.9457 78.5393
Item Value Threshold Converged?
Maximum Force 0.000048 0.000450 YES
RMS Force 0.000015 0.000300 YES
Maximum Displacement 0.001464 0.001800 YES
RMS Displacement 0.000541 0.001200 YES
Predicted change in Energy=-4.001091D-08
Optimization completed.
-- Stationary point found.
Main peaks (Frequency/cm-1): 420, 582
Disssociation Energy of trans isomer
E(monomer) = -1176.19013697 a.u.
E(adduct) = -2352.41629858 a.u
ΔE = E(adduct)-[2x(monomer)]
ΔE = -2352.41629858 - -(2x-1176.19013697)
ΔE = -0.03602464 a.u.
1 a.u. = 2625.5 kJ mol-1
Enthalpy of dissociation = -0.03602464 a.u. * 2625.5 = -94.6 kJ mol-1
Relative Energies
| Molecule | Energy (a.u.) | Energy (kJ/mol) | Relative Energy (kJ/mol) |
|---|---|---|---|
| Cl2Al(μ-Br2)AlCl2 | -2352.40630798 | -6176242.762 | 26.23 |
| BrClAl(μ-Br,Cl)AlCl2 | -2352.41626677 | -6176268.908 | 0.08 |
| Trans-BrClAl(μ-Cl2)AlClBr | -2352.41629858 | -6176268.992 | 0.00 |
| Cis-BrClAl(μ-Cl2)AlClBr | -2352.41109944 | -6176255.342 | 13.65 |
Trans-BrClAl(μ-Cl2)AlClBr has the lowest energy and so is the most stable isomer. This is because when broken into the two constituent monomers, there is an electron acceptor (acting as the lewis acid) and an electron donor (acting as a lewis base)in each of the fragments. The trans isomer was therefore assigned an energy of zero, so that the relative energies of the other isomers could be more easily compared to this.
The Cl2Al(μ-Br2)AlCl2 isomer has the highest relative energy and so is the least stable. This is because Br has more diffuse orbitals than Cl, so the orbital overlap of the Al atoms with the bridging Br atoms is less effective than with Cl atoms.
Monomer
Optimisation and pseudo-potential
| AlBrCl2 | ||
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | GEN | |
| Final Energy | -1176.19013697 a.u. | |
| Gradient | 0.00000291 a.u. | |
| Dipole Moment | 0.1134 Debye | |
| Point Group | C2v | |
| CPU Time | 46.6 seconds | |
Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000002 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000012 0.001200 YES
Predicted change in Energy=-1.397817D-10
Optimization completed.
-- Stationary point found.
Frequency Analysis
Low frequencies --- -2.4223 -0.0067 -0.0065 -0.0059 2.7464 2.9629 Low frequencies --- 120.5194 133.8347 185.7791
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.000021 0.001800 YES
RMS Displacement 0.000009 0.001200 YES
Predicted change in Energy=-1.460239D-10
Optimization completed.
-- Stationary point found.
1.↑ M. Schuurman, W. Allen, H. Schaefer, Journal of Computational Chemistry, 2005, 26, 1106
2.↑ J. Blixt et al., J. Am. Chem. Soc. , 117, 1995, pp 5089 - 5104
3.↑ Bondi (1964) J. Phys. Chem. 68, 441
4.↑ Paula, Peter Atkins, Julio de (2009). Elements of physical chemistry (5th ed. ed.). Oxford: Oxford U.P. pp. 459. ISBN 978-0-19-922672-6.