Rep:Mod:ilikecake
Computational Lab Module 1
This project aims to use computational modelling methods to predict the geometry and regioselectivity of the hydrogenation of a cyclopentene dimer, and the conformation / atropisomerism of a large ketone intermediate in the synthesis of taxol; to then use semi-empirical and DFT molecular orbital theory to investigate the origins of the regioselectivity of the electrophilic carbenylation of a chloro-substituted bicyclic diene and monosaccharide chemistry and the mechanism of glycosidation. In the second part of the project aims to evaluate a reported literature molecule against spectra obtained from the computational software to judge if the molecule has actually been found.
Dimerisation of cyclopentene
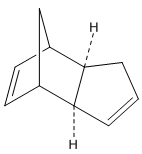
Cyclopentene readily undergoes Diels-Alder reaction with itself at room temperature to produce a dimer, which can take either endo or exo forms. Here we shall compare the various data obtained from modelling operations to judge if the reaction proceeds by and is under thermodynamic or kinetic control.
The exo cyclopentene dimer was created using ChemBio3D, and its geometry optimised using the MM2 force field option. We see a very positive bend contribution, indicating the predicted bonds are bent far from the natural angle.
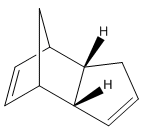
The endo cyclopentene dimer was created using ChemBio3D, and its geometry optimised using the MM2 force field option.
| endo | exo | ||||||
|---|---|---|---|---|---|---|---|
|
| ||||||
------------MM2 Minimization------------ Note: All parameters used are finalized (Quality = 4). Stretch: 1.2507 Bend: 20.8476 Stretch-Bend: -0.8358 Torsion: 9.5109 Non-1,4 VDW: -1.5430 1,4 VDW: 4.3195 Dipole/Dipole: 0.4476 Total Energy: 33.9975 kcal/mol Calculation completed ------------------------------------ |
------------MM2 Minimization------------ Note: All parameters used are finalized (Quality = 4). Stretch: 1.2855 Bend: 20.5797 Stretch-Bend: -0.8383 Torsion: 7.6561 Non-1,4 VDW: -1.4172 1,4 VDW: 4.2332 Dipole/Dipole: 0.3775 Total Energy: 31.8765 kcal/mol Calculation completed ------------------------------------ | ||||||
The greatest difference between the minimised structures is seen in the torsion energies.
Comparing the relative energies of the endo and exo forms, it is seen that the exo form is lower by 2.121 kcal/mol. Yet cyclopentadiene dimerises to form specifically the endo dimer. This must indicate therefore that the reaction is under kinetic rather than thermodynamic control since the least stable isomer is formed.
This selectivity can be explained by transition states in the Diels-Alder reaction, having in the endo form a stabilisation of ion charges in resonance that are not found in the exo form; and favourable p orbital HOMO-LUMO interactions on approach of one cyclyopentene on the other leading to the endo form, again which are not found in production of the exo form. These energetically favourable interactions result in almost complete production of the endo form.
Hydrogenation of cyclopentene dimer
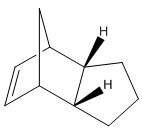
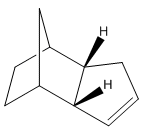
Looking specifically at the endo dimer, its hydrogenation can yield either 3 or 4 below:
| dimer 3 | dimer 4 | ||||||
|---|---|---|---|---|---|---|---|
|
| ||||||
------------MM2 Minimization------------ Note: All parameters used are finalized (Quality = 4). Stretch: 1.2346 Bend: 18.9413 Stretch-Bend: -0.7612 Torsion: 12.1224 Non-1,4 VDW: -1.5021 1,4 VDW: 5.7286 Dipole/Dipole: 0.1631 Total Energy: 35.9266 kcal/mol Calculation completed ------------------------------------ |
------------MM2 Minimization------------ Note: All parameters used are finalized (Quality = 4). Stretch: 1.0963 Bend: 14.5254 Stretch-Bend: -0.5493 Torsion: 12.4978 Non-1,4 VDW: -1.0709 1,4 VDW: 4.5120 Dipole/Dipole: 0.1406 Total Energy: 31.1520 kcal/mol Calculation completed ------------------------------------ | ||||||
The hydrogenated dimers 3 and 4 were created in ChemBio3D and optimised.
By comparing the relative energies of each form, it can be deduced which is the most stable and hence which double bond has the greater ease of hydrogenation.
It can be seen that dimer 4 has the lower energy, by 4.7746 kcal/mol. A possible explanation for this is that in conformer 3, there is greater alkene strain as it is close to bridging carbon.
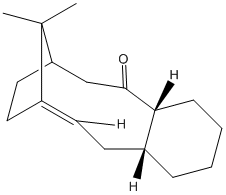
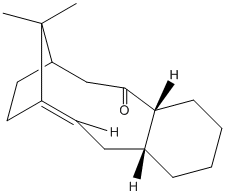
An intermediate in the synthesis of taxol can be formed with either the carbonyl group facing upwards or downwards relative to the ring plane.This particular intermediate is formed from an oxy-cope rearrangement.
We can compare the relative energies of each atropisomer to deduce each relative stability:
| carbonyl up | carbonyl down | ||||||
|---|---|---|---|---|---|---|---|
|
| ||||||
------------MM2 Minimization------------ Warning: Some parameters are guessed (Quality = 1). Stretch: 6.4271 Bend: 89.9942 Stretch-Bend: -0.3927 Torsion: 19.2166 Non-1,4 VDW: 2.6660 1,4 VDW: 29.8332 Dipole/Dipole: -2.7872 Total Energy: 144.9571 kcal/mol Calculation completed ------------------------------------ |
------------MM2 Minimization------------ Separating coincident atoms: Lp(45)-Lp(46) Warning: Some parameters are guessed (Quality = 1). Stretch: 2.7329 Bend: 13.8241 Stretch-Bend: 0.3477 Torsion: 20.0140 Non-1,4 VDW: -0.8732 1,4 VDW: 13.7357 Dipole/Dipole: -1.5753 Total Energy: 48.2059 kcal/mol Calculation completed ------------------------------------ | ||||||
| carbonyl up | carbonyl down | ||||||
|---|---|---|---|---|---|---|---|
|
| ||||||
------------MMFF94 Minimization------------ Final Energy: 81.3234 kcal/mol Calculation completed ------------------------------------ |
------------MMFF94 Minimization------------ Final Energy: 66.6176 kcal/mol Calculation completed ------------------------------------ | ||||||
MM2 optimisation was carried out, and it can be seen that the energy of the molecule with the carbonyl facing downwards is far lower in energy than the atropisomer with carbonyl facing upwards.
The energy optimisation calculation was repeated using the MMFF94 force-field
This essentially means that the carbonyl up-facing conformer is more reactive. This can be explained by consideration of olefinic strain, that is where an alkene is connected to a bridgehead carbon as is the case in this molecule. Alkenes will usually attempt to avoid such a placement (Bredt's Rule describes this, the alkene behaves more like a single bond, as a result of steric strain). The olefinic strain energy is calculated by subtracting the strain energy of the parent alkane from the strain energy of the alkene. Having said that alkenes will usually attempt to avoid such a placement (in molecules with positive olefinic strain energy), 'hyperstable olefins' exist that possess negative olefenic strain energies, ie, are less strained than their parent alkane, with this molecule being an example.
Modelling Using Semi-empirical Molecular Orbital Theory
Regioselective Addition of Dichlorocarbene to a diene
The reaction between dichlorocarbene and 9-chloromethanonapthelene will be studied, by minimising the diene by MM2 and Mopac; comparing the differences between the optimised method structures, and then examining the relevant molecular orbitals and electropotential surfaces in order to explain the high pi selectivity in this reaction.
------------MM2 Minimization------------ Warning: Some parameters are guessed (Quality = 1). Iteration 147: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 0.6195 Bend: 4.7370 Stretch-Bend: 0.0401 Torsion: 7.6591 Non-1,4 VDW: -1.0672 1,4 VDW: 5.7937 Dipole/Dipole: 0.1123 Total Energy: 17.8945 kcal/mol Calculation completed ------------------------------------
------------ Mopac Interface ------------ Model: molecule 12.mol Mopac Job: AUX RM1 CHARGE=0 EF GNORM=0.100 SHIFT=80 Finished @ RMS Gradient = 0.07818 (< 0.10000) Heat of Formation = 22.82766 Kcal/Mol -----------------------------------------
Distance between pair : 4.8A
The MM2 and MOPAC optimised structures were overlayed, tethered by four atoms, to account for any differences in the structure. The correlation is actually very good, with largest discrepancy between the C-Cl bond.
Overlay |
Molecular Orbital Analysis
------------ Gaussian Interface ------------ Model: Untitled-2 1) Gaussian Job: # B3LYP/6-31G Test Charges (Electron Density): C(1) 4.931858 C(2) 4.962210 C(3) 5.128303 C(4) 5.499631 C(5) 5.499893 C(6) 5.141252 C(7) 5.128165 C(8) 4.962360 C(9) 4.931958 C(10) 5.141565 C(11) 5.958556 Cl(12) 16.935590 H(13) 0.609551 H(14) 0.599577 H(15) 0.609277 H(16) 0.620130 H(17) 0.607259 H(18) 0.613774 H(19) 0.609249 H(20) 0.620130 H(21) 0.599590 H(22) 0.609570 H(23) 0.607223 H(24) 0.613615 H(25) 0.522187 Charges (Mulliken Charges): C(1) -0.085071 C(2) -0.106997 C(3) -0.296967 C(4) 0.063118 C(5) 0.063091 C(6) -0.306604 C(7) -0.297004 C(8) -0.107073 C(9) -0.084984 C(10) -0.306862 C(11) -0.387743 Cl(12) 0.044551 H(13) 0.112316 H(14) 0.120321 H(15) 0.147722 H(16) 0.136015 H(17) 0.144824 H(18) 0.145632 H(19) 0.147761 H(20) 0.136017 H(21) 0.120325 H(22) 0.112320 H(23) 0.144765 H(24) 0.145840 H(25) 0.194688 Gaussian Interface: Dipole = (1.9030, 0.0017, -1.4691) 2.4041 Debye Gaussian Interface: SCF Energy = -556594.18 Kcal/Mol --------------------------------------------
MO alkenes |
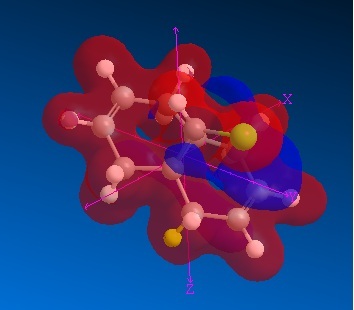
Electrostatic potential
We know that the electrophilic addition occurs endo- to the bridged chlorine atom, and the potential diagram clearly shows the difference in electron density, with the greater density on the endo-pi bond.
The HOMO-LUMO region is important in understanding why the reaction shows such regiospecificity. Below is a visual summary of some of the key molecular orbitals:
| MO | Image | Energy (eV) |
|---|---|---|
| LUMO +2 | 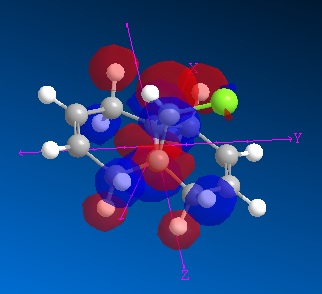
|
15.742 |
| LUMO +1 | 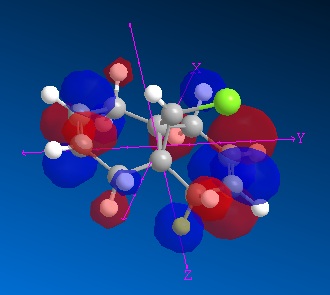
|
1.339 |
| LUMO | 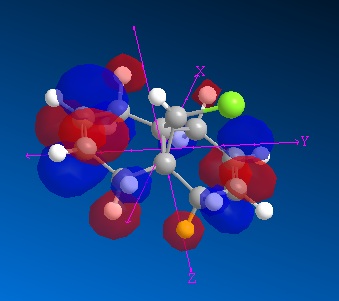
|
1.008 |
| HOMO | 
|
-10.497 |
| HOMO -1 | 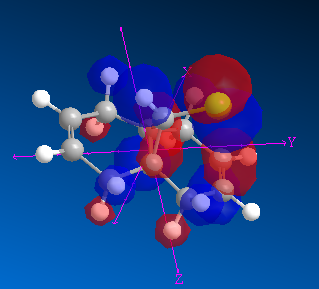
|
-10.916 |
| HOMO -2 | 
|
-13.620 |
An important aspect of these MOs is the interaction between the HOMO-1 and LUMO+2 which together combine to form an anti-periplanar arrangement that leads to an increase in the bonding character of the C-Cl antibonding sigma orbital, and an increase in electron density on the exo pi bond, into the antibonding orbitals (weakening of this bond).
Stretching frequency analysis
| vibration | energy | intensity |
|---|---|---|
|
770.80 | 25.1517 |
|
1737.04 | 4.1982 |
|
1757.35 | 3.9362 |
It is seen that the exo-pi bond stretching frequency occurs at 1737.04 compared to 1757.35 for the endo-pi bond; indicating that the exo-pi bond has been weakened and the alkene character subsequently reduced, as explained by the molecular orbitals.
The most intense C-Cl stretch is shown.
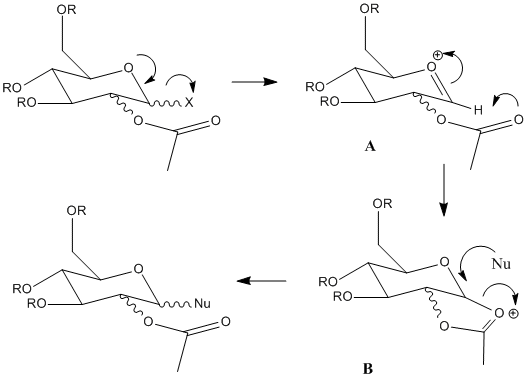
Monosaccharide chemistry and the mechanism of glycosidation
This reaction is studied as it is highly stereospecific; we are able to use molecular modelling to compare OAc conformers of the intermediates A and B.
Four different conformers can be created by the ester being upwards or downwards relative to the ring plane, and for each of these conformations the acetyl can be upwards facing or downwards facing.
| MM2 | MMFF94 | ||||||
|---|---|---|---|---|---|---|---|
| 1 (u, u) | 2 (u, d) | 3 (d, u) | 4 (d, d) | 1 (u, u) | 2 (u, d) | 3 (d, u) | 4 (d, d) |
| 33.2031 kcal/mol | 33.0305 kcal/mol | 33.4948 kcal/mol | 29.6599 kcal/mol | 50.23459 Kcal/Mol | 21.20939 Kcal/Mol | 21.04950 Kcal/Mol | 65.28947 Kcal/Mol |
| MM2 | MMFF94 | ||||||
|---|---|---|---|---|---|---|---|
| 1 (u, u) | 2 (u, d) | 3 (d, u) | 4 (d, d) | 1 (u, u) | 2 (u, d) | 3 (d, u) | 4 (d, d) |
| 30.1140 kcal/mol | 32.8872 kcal/mol | 36.0766 kcal/mol | 37.1961 kcal/mol | 15.96182 Kcal/Mol | 15.92375 Kcal/Mol | 12.42602 Kcal/Mol | 12.44866 Kcal/Mol |
It is observed that molecule A has lower energy than molecule B in the conformers in which the ester C-O is pointing downwards. In molecule B, there exists high conformational strain, and the distinct up-down conformers are not readily distinguished as a result of the formation of the heterocycle ring.
The Mopac energies are lower as the technique is able to reorganise and form new bonds (MM2 can only consider the bonds that have been entered)
Selected Mopac and MM2 optimaztions have been overlayed to visually observe the differences in optimised structure between the methods:
Overlays
A u, u
A u, u |
A d, u
A d, u |
B u, u
B u, u |
B d, u
B d, u |
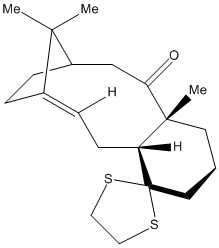
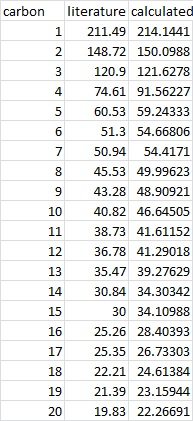
The 13C NMR of the below intermediate in taxol synthesis has been calculated and compared to literature.
It was optimised with DFT=mpw1pw91 using basis set of 6-31G with d, p polarization.
------------MM2 Minimization------------ Warning: Some parameters are guessed (Quality = 1). Iteration 2: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 5.3598 Bend: 20.3276 Stretch-Bend: 0.8745 Torsion: 22.5051 Non-1,4 VDW: -0.3087 1,4 VDW: 17.3697 Dipole/Dipole: -1.7587 Total Energy: 64.3693 kcal/mol Calculation completed ------------------------------------
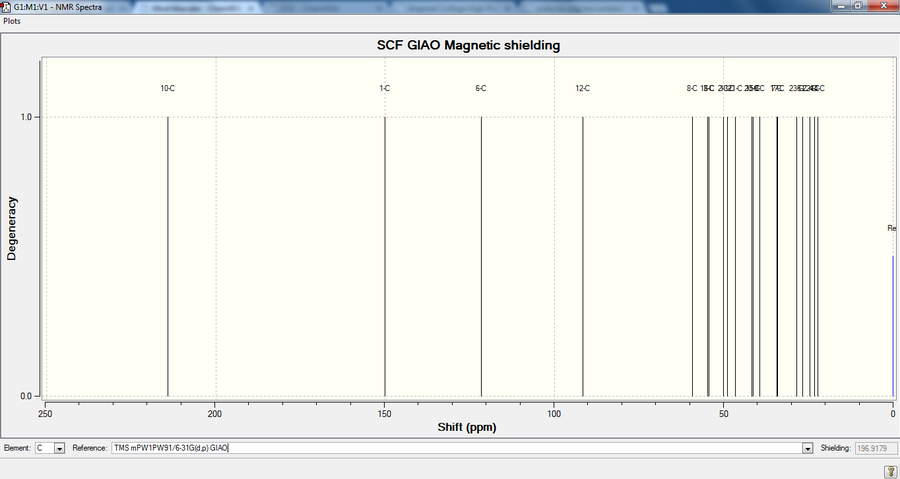
The NMR was referenced to TMS mPW1PW91/6-31Gd,p GIAO.
There is overall good agreement with the literature data; the greatest difference in chemical shift is for carbon 4, likely as a result of spin-orbit coupling, as this carbon is adjacent to the dithiane group.
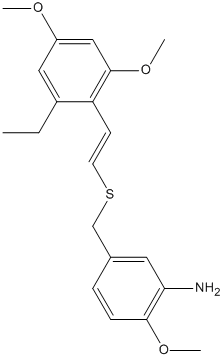
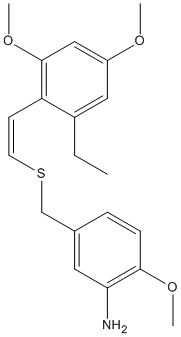
E / Z isomers of 2',4',6'-trimethoxystyryl-4-methoxy-3-aminobenzyl sulfide
Preparation of E-vinyl sulfides is well known, but it has been challenging to produce Z-vinyl sulfides. A method has been reported for stereoselective Z-styryl benzyl sulfides via the hydrothiolation of aryl acetylenes by nenzyl thiols with a radical initiator.
One such pair of reported E/Z isomers will be modelled using ChemBio3D, and their 13C and 1H spectra predicted, and compared to the literature data.
E isomer (Molecule 18)
Molecule 18 |
------------MM2 Minimization------------ Pi System: 1 7 8 9 11 10 19 Pi System: 14 13 3 4 5 12 6 15 16 Stretch: 2.6363 Bend: 10.5317 Stretch-Bend: 0.2309 Torsion: -8.3910 Non-1,4 VDW: 0.9518 1,4 VDW: 24.0105 Dipole/Dipole: -0.1690 Total Energy: 29.8011 kcal/mol Calculation completed ------------------------------------
Z isomer (Molecule 17)
------------MM2 Minimization------------ Pi System: 1 7 8 9 11 10 19 Pi System: 14 13 3 4 5 12 6 15 16 Warning: Some parameters are guessed (Quality = 1). Iteration 2855: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 2.6862 Bend: 12.5488 Stretch-Bend: 0.3485 Torsion: -14.0614 Non-1,4 VDW: -0.8023 1,4 VDW: 23.8250 Dipole/Dipole: -0.7128 Total Energy: 23.8319 kcal/mol Calculation completed ------------------------------------
Molecule 17 |
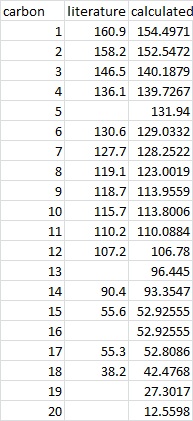
13C NMR
The NMR was run with DFT = mpw1pw91 using a 6-31G (d,p) polarization basis set.
Both NMR were referenced to TMS mPW1PW91/6-31Gd,p CDCl3 GIAO.
There is overall excellent correlation with the literature, with most values within a 5ppm range. There is greatest difference for carbon 1.
As with the Z isomer, the E isomer shows very good correlation between the predicted spectra and the literature.
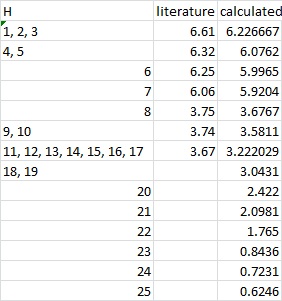
1H NMR
Both NMR was referenced to TMS mPW1PW91/6-31Gd,p CDCl3 GIAO.
Prediction of the nJ-J H couplings
The H coupling constants have been predicted, and the log files are available on the D-space links below:
mol 17
[D-space]
mol 18
[D-space]
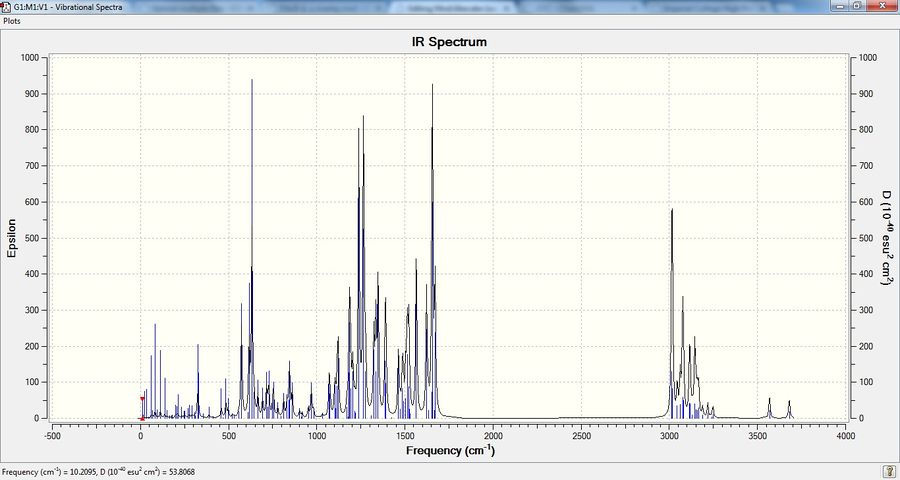
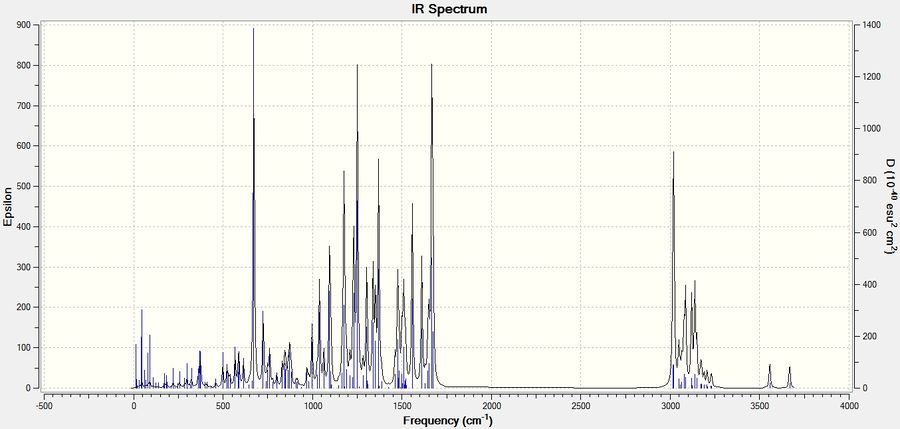
Prediction of IR Spectrum
molecule 17
Molecule 18
Molecule 17
Sum of electronic and thermal Free Energies= -1455.428366 (Hartrees), -913281.299665 kcal/mol
molecule 18
Sum of electronic and thermal Free Energies= -1455.428503 (Hartrees), -913281.385633 kcal/mol
Hence, the difference in free energy between the isomers is 0.085967 kcal/mol.
Summary
The 13C and 1H NMR have been predicted and compared to the reported values in literature. Only the NMR data were available for these compounds, it would have been useful to compare the IR spectra and develop further data. Nonetheless the predicted spectra may be useful for future reference. It is observed that there is good correlation between the predicted nmr data and literature, indicating that the modelling process is a useful tool for predicting the spectra, and that the literature compounds reported are likely correctly identified.
References
taxol intermediate spectra data - http://pubs.acs.org/doi/pdf/10.1021/ja00157a043
isomers - http://pubs.rsc.org/en/content/articlepdf/2013/ob/c3ob27220f