Rep:Mod:huyi
NH3
Bond angle of the H-N-H
105.74115°
Bond length of the H-N bond
1.01798 Å (Literature value: 101 pm[1])
This value is closed to the literature and small amount of difference is negligible.
Key information on NH3
Molecule Name NH3 Calculation Type FREQ Calculation Method RB3LYP Basis Set 6-31G(d,p) Charge 0 Spin Singlet E(RB3LYP) -56.55776873 a.u. RMS Gradient Norm 0.00000485 a.u. Imaginary Freq 0 Dipole Moment 1.8466 Debye Point Group C3V
"Item" section in the log file of NH3
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986262D-10
Optimization completed.
-- Stationary point found.
A Jmol dynamic image of NH3
NH3 optimisation |
Download link:File:QIUNILE.LOG
Questions
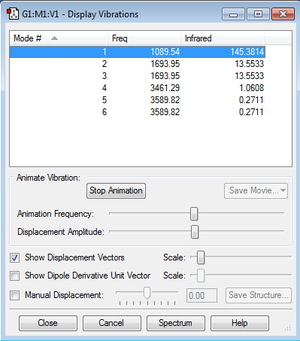
How many modes do you expect from the 3N-6 rule?
6 modes as shown in the frequency analysis table.
which modes are degenerate (ie have the same energy)?
the 2rd and the 3rd, the 5th and the 6th as those two pairs have the similar frequency hence similar energy.
which modes are "bending" vibrations and which are "bond stretch" vibrations?
Bending vibrations : Mode 1,2,3 because their frequencies are in the range of 1000-1600.
Stretching Vibrations: Mod 4,5,6 because their frequencies are approximately 3500.
which mode is highly symmetric?
Mode 4 as it has three symmetric planar.
one mode is known as the "umbrella" mode, which one is this?
Mode 1 due to "opening-umbrella motion".
how many bands would you expect to see in an experimental spectrum of gaseous ammonia?
2 bands. The rest of bands would be too small to be observed in an experimental spectrum.
The charge on the N-atom and H-atoms.
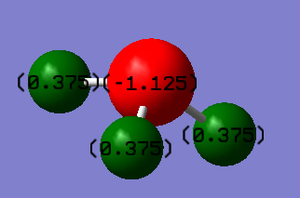
N:-1.125 H:0.375
Negative charge is expected found on N atom and positive charges is expected found on H atoms.
As Nitrogen is a element with higher electronegativity than hydrogen, therefore electrons will be dragged further from hydrogen atoms.
N2
Bond length of the N-N bond
1.10550 Å (Literature value: 1.10 pm[1])
This optimized value is closed and small amount of difference is negligible.
Key information on n2
Molecule Name N2 Calculation Type FREQ Calculation Method RB3LYP Basis Set 6-31G(D,P) Charge 0 Spin Singlet Total Energy -109.52412868 a.u. RMS Gradient Norm 0.00000060 a.u. Imaginary Freq Dipole Moment 0.0000 Debye Point Group D∞h
"Item" section in the log file of H2
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.401082D-13
Optimization completed.
-- Stationary point found.
A Jmol dynamic image of N2
N2 optimisation |
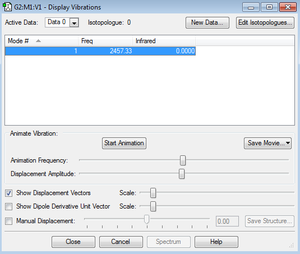
Frequency & Charges
2457.33, N2 is a linear molecule therefore due to 3N-5 rule only one mode would be observed.
Charges on both atoms are 0 because they have same electronegativity.
Download link:File:DANQI.LOG
H2
Bond length of the N-N bond
0.74279 Å (literature value: 74 pm[1])
The optimized value is consistent with the literature value.
Key information on H2
Molecule Name H2 Calculation Type FREQ Calculation Method RB3LYP Basis Set 6-31G(D,P) Charge 0 Spin Singlet Total Energy -1.17853936 a.u. RMS Gradient Norm 0.00000017 a.u. Imaginary Freq Dipole Moment 0.0000 Debye Point Group D∞h
"Item" section in the log file of H2
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
A Jmol dynamic image of N2
H2 optimisation |
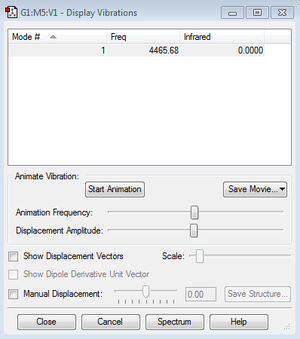
Frequency & Charges
4465.68, H2 is a linear molecule therefore due to 3N-5 rule only one band would be observed.
Charges on both atoms are 0 because they have same electronegativity.
Download link:File:QINGQIDEZUIJIAHUA.LOG
Haber-Bosch process energy calculation
E(NH3)= -56.55776873 a.u. = -148492.421801 kJ/mol
2*E(NH3)= -113.11553746 a.u. = -296984.843601 kJ/mol
E(N2)= -109.52412868 a.u. = -287555.599849 kJ/mol
E(H2)= -1.17853936 a.u. = -3094.25508968 kJ/mol
3*E(H2)= -3.53561808 a.u. = -9282.76526904 kJ/mol
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.0557907 a.u. = -146.47848285 kJ/mol
ΔE is a negative value hence this reaction is exothermic reaction and the product of the reaction is more stable.
Choice of small molecule Cl2
Bond length of the H-N bond
2.04174 Å (Literature value 1.99 pm)
This value is more different from the literature value than other elements. Possible explanation is the software used different method of optimization from the realistic condition or different methods of optimization should be applied to different elements.
Key information on Cl2
Molecule Name Cl2 Calculation Type FREQ Calculation Method RB3LYP Basis Set 6-31G(d,p) Charge 0 Spin Singlet E(RB3LYP) -920.34987886 a.u. RMS Gradient Norm 0.00002511 a.u. Imaginary Freq 0 Dipole Moment 0.0000 Debye Point Group D*H
"Item" section in the log file of Cl2
Item Value Threshold Converged? Maximum Force 0.000043 0.000450 YES RMS Force 0.000043 0.000300 YES Maximum Displacement 0.000121 0.001800 YES RMS Displacement 0.000172 0.001200 YES Predicted change in Energy=-5.277285D-09 Optimization completed. -- Stationary point found.
A Jmol dynamic image of Cl2
Cl2 optimisation |
Download link:File:LVQI +1.LOG
Frequency & Charges
520.32, Cl2 is a linear molecule therefore due to 3N-5 rule only one band would be observed.
Charges on both atoms are 0 because they have same electronegativity.

Comments on the MOs
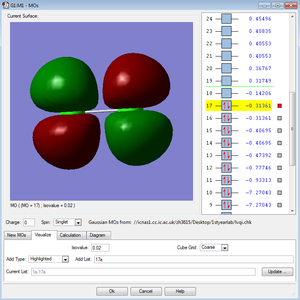
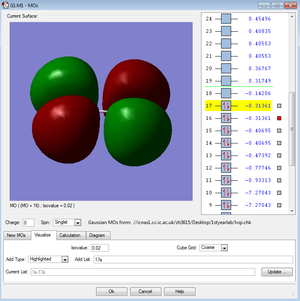
←Description of MO1 and MO2:
These two molecular orbitals of the Cl2 molecule have the highest energy hence they are in the HOMO or LUMO region. They are the occupied and antibonding combinations of the 3Px and 3Py orbitals, which are the anitbonding pi orbitals.
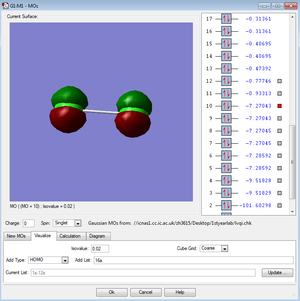
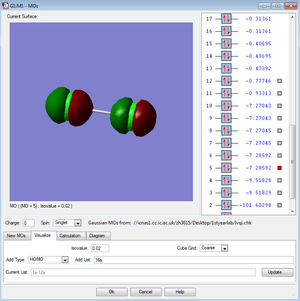
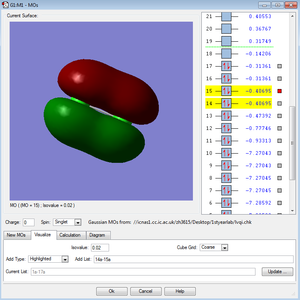
←Description of MO3 and MO4:
These two molecular orbitals of the Cl2 molecule made of the 2Px and 2Pz atomic orbitals. Molecular orbitals of the combinations of the 2Pxs and 2Pys have the same energy and they are both antibonding orbitals. Combination of the 2Pzs has lower energy in the MO diagram due to the interaction between the AOs. They are all occupied and not in the HOMO/LUMO region.
←Description of MO5:
This Molecular orbital is the pi orbital formed by either of 3Px or 3Py. It has lower energy than the HOMO/LUMO region and it is occupied. It is an bonding orbital as shown in the screenshot.
Reference
[1]Huheey, pps. A-21 to A-34; T.L. Cottrell, "The Strengths of Chemical Bonds," 2nd ed., Butterworths, London, 1958; B. deB. Darwent, "National Standard Reference Data Series," National Bureau of Standards, No. 31, Washington, DC, 1970; S.W. Benson, J. Chem. Educ., 42, 502 (1965).
