Rep:Mod:hsh16
Ammonia NH3
Gaussian Optimization Data
Calculation Method = RB3LYP
Basis Set = 6-31G(d,p)
E(RB3LYP) = -56.55776873 a.u.
RMS Gradient Norm = 0.00000485 a.u.
Point Group = C3V
Optimised N-H bond distance = 1.01798 Angstrom
Optimised H-N-H bond angle = 105.741 degrees
Gaussian optimization file here.
Charge distribution: -1.125 (N) and 0.375 (H). Matches expectations as nitrogen is the more electronegative atom and is therefore able to draw negatively charged electron density onto itself.
Optimization Confirmation Text (From Log File)
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986281D-10
Optimization completed.
-- Stationary point found.
Questions
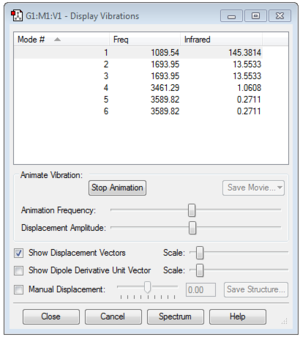
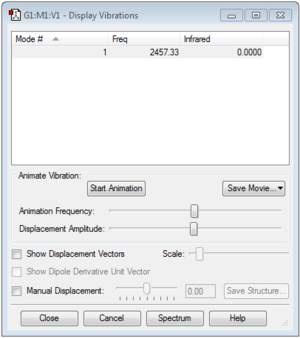
1. How many modes do you expect from the 3N-6 rule?
6 modes are expected.
2. Which modes are degenerate (ie have the same energy)?
Modes 2 & 3 are degenerate, as are 5 & 6.
3. Which modes are "bending" vibrations and which are "bond stretch" vibrations?
1,2 and 3 are bending vibrations while 4, 5 and 6 are bond stretch vibrations.
4. Which mode is highly symmetric?
Mode 4.
5. One mode is known as the "umbrella" mode, which one is this?
Mode 1.
6. How many bands would you expect to see in an experimental spectrum of gaseous ammonia?
3 bands are to be expected from the spectrum.
Hydrogen H2 & Nitrogen N2
Hydrogen Gaussian Optimisation Data
Calculation Method = RB3LYP
Basis Set = 6-31G(d,p)
E(RB3LYP) = -1.17853936 a.u.
RMS Gradient Norm = 0.00000017 a.u.
Point Group = D*H
Gaussian optimization file here.
Optimisation Confirmation Text (From Log File)
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
Nitrogen Gaussian Optimization Data
Calculation Method = RB3LYP
Basis Set = 6-31G(d,p)
E(RB3LYP) = -109.52412868 a.u.
RMS Gradient Norm = 0.00000060 a.u.
Point Group = D*H
Gaussian optimization file here.
Optimisation Confirmation Text (From Log File)
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.400954D-13
Optimization completed.
-- Stationary point found.
Energy Calculations
E(NH3) = -56.55776873 a.u.
2*E(NH3) = -113.11553746 a.u.
E(N2) = -109.52412868 a.u.
E(H2) = -1.17853936 a.u.
3*E(H2) = -3.53561808 a.u.
ΔE = 2*E(NH3) - [E(N2)+3*E(H2)] = -0.0557907 a.u. = -146.48 kJ
Molar ΔE = -73.24 kJ mol-1
Literature Value = -33.79 kJ mol-1[1]
- ↑ 1.C. Vanderzee and D. King, The Journal of Chemical Thermodynamics, 1972, 4, 675-683.
Calculation deviates from the literature value, difference results from the fact that the calculations may approximate some aspects of the molecule.
ΔE is negative, therefore the ammonia product is more stable.
Chosen Molecule: Silanone SiOH2
Gaussian Optimisation Data
Calculation Method = RB3LYP
Basis Set = 6-31G(D,P)
Total Energy = -365.90001403 a.u.
RMS Gradient Norm = 0.00000941 a.u.
Point Group = C1
Charge Distribution: 1.472(S), -0.236(H), -1.001(O)
Bond Angles: H-Si-O = 124.156 degrees, H-Si-H = 111.686 degrees
Gaussian optimization file here.
Optimisation Confirmation Text (From Log File)
Item Value Threshold Converged?
Maximum Force 0.000023 0.000450 YES
RMS Force 0.000009 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000017 0.001200 YES
Predicted change in Energy=-5.109767D-10
Optimization completed.
-- Stationary point found.
Interesting Molecular Orbitals

The second molecular orbital in the molecule. Is an antibonding orbital formed by the out-of-phase overlap of the 1s orbitals of the atoms, with the greatest contribution coming from the oxygen atom. Very deep in energy compared to other occupied orbitals at -19.92kJ mol-1, although not the deepest orbital present.

The third molecular orbital in the molecule. It is a bonding orbital formed by the in-phase overlap of the 2s orbitals of the atoms with the greatest contribution coming from the silicon atom. Much higher in compared to the first bonding-antibonding pair of orbitals but still not the highest in energy in the molecule.

The eleventh molecular orbital in the molecule. It is a bonding orbital formed by the in-phase overlap of the 3p orbitals of the atoms. Its energy is in the HOMO region, being the second-highest occupied orbital with regards to energy.

The twelfth molecular orbital in the molecule. It is an antibonding orbital formed by the out-of-phase overlap of the 3p orbitals of the atoms. It is the molecule's HOMO, at -0.28kJ mol-1 and is also the molecule's largest occupied molecular orbital.

The thirteenth molecular orbital. This is the molecule's LUMO and is the first unoccupied orbital encountered in the molecule. The greatest contribution comes from the silicon atom.
Additional Investigation: Fluorine F2
Gaussian Optimisation Data
Calculation Method = RB3LYP
E(RB3LYP) = 6-31G(D,P)
Total Energy = -199.49825218 a.u.
RMS Gradient Norm = 0.00007365 a.u.
Point Group = D*h
Gaussian optimization file here.
Optimisation Confirmation Text (From Log File)
Item Value Threshold Converged?
Maximum Force 0.000128 0.000450 YES
RMS Force 0.000128 0.000300 YES
Maximum Displacement 0.000156 0.001800 YES
RMS Displacement 0.000221 0.001200 YES
Predicted change in Energy=-1.995024D-08
Optimization completed.
-- Stationary point found.
