Rep:Mod:haien910
NH3
test molecule |
NH3 is a colorless gas with a characteristic pungent smell. Fritz Haber and Carl Bosch developed the Haber–Bosch process in 1909 to produce ammonia from the nitrogen in the air.
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d.p) |
| Final Energy | -56.55776873 a.u., -148492.43 kJ/mol |
| RMS Gradient Norm | 0.00000485 a.u. |
| Point Group | C3V |
Optimised N-H bond distance = 1.01798 Å
Optimised H-N-H bond angle = 105.741°
Optimisation Table
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986281D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7412 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7412 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7412 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8571 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Vibrational Modes of NH3
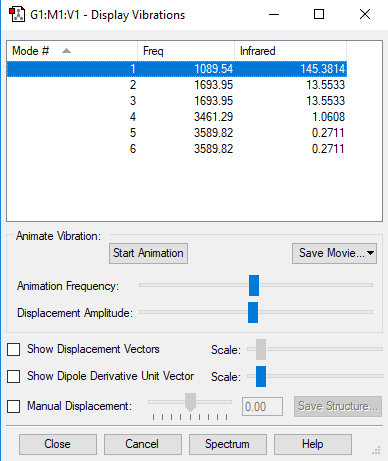
Since NH3 is a non-linear molecule, using the rule 3N-6 where N = 4, we would expect it to have 6 vibrational modes. This is correctly predicted by the Gaussview software.
Degenerate orbitals are orbitals that have the same energy. Modes 2 & 3 are degenerate to each other whereas 5 & 6 are degenerate to each other. This is deduced by looking at their frequencies; if they're degenerate, they would have equivalent frequencies. Modes 1, 2 and 3 are bending modes whereas 4, 5 and 6 are stretching modes.
Mode 4 is highly symmetric and Mode 1 is known as the 'umbrella' mode.
Charge distribution on N = - 1.125
Charge distribution on H = + 0.375
N is more electronegative than H, it has a higher tendency to attract a bonding pair of electrons and so there is a partial negative charge on N atom and a partial positive charge on H atom. The magnitude of charge of N atom about 3 times greater than of H atom because there is only 1 N for every 3 H.
IR Spectrum of NH3
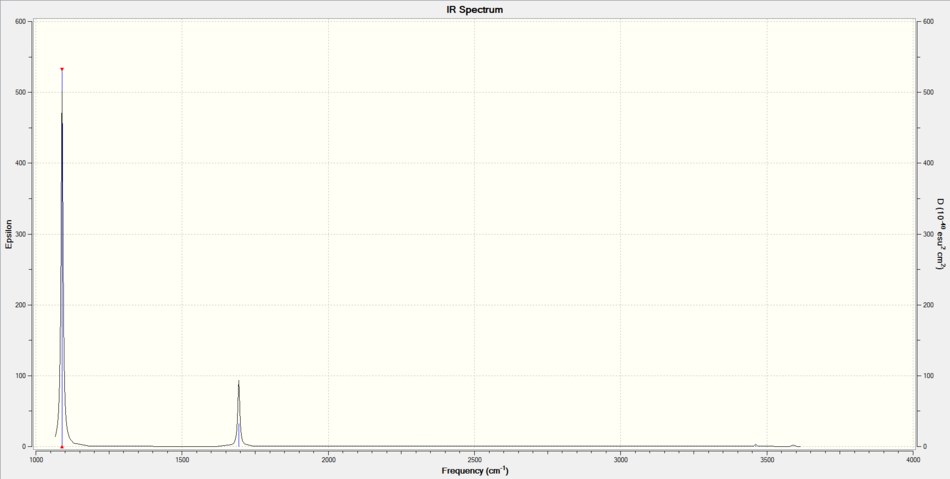
2 peaks were observed and this is due to the 2 degenerate pairs (2 & 3 and 5 & 6). Mode 4 is a symmetric stretch, it has no change in dipole moment therefore is IR inactive. It is worth noting that intramolecular interaction like hydrogen bonding between NH3 was not detected by Gaussview.
H2
test molecule |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d.p) |
| Final Energy | -1.17853936 a.u., -3094.26 kJ/mol |
| RMS Gradient Norm | 0.00000017 a.u. |
| Point Group | D∞H |
| Bond Length | 0.74279 Å |
H2 does have any charge distribution because there is no difference in electronegativity values.
Optimisation Table
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 0.7428 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Vibrational Modes of H2
H2 is a linear molecule, using the rule 3N-5 where N = 2, we would expect it to only have 1 vibrational mode.
IR Spectrum of H2
H2 has no net change in dipole moment, so it is IR inactive, therefore no peaks were observed on IR spectrum.
N2
test molecule |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d.p) |
| Final Energy | -109.52412868 a.u., -287555.62 kJ/mol |
| RMS Gradient Norm | 0.00000060 a.u. |
| Point Group | D∞H |
| Bond Length | 1.10550 Å |
N2 does have any charge distribution because there is no difference in electronegativity values.
Optimisation Table
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.401020D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1055 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Vibrational Modes of N2
N2 is a linear molecule, using the rule 3N-5 where N = 2, we would expect it to only have 1 vibrational mode.
IR Spectrum of H2
N2 has no net change in dipole moment, so it is IR inactive, therefore no peaks were observed on IR spectrum.
Thermodynamics of Haber Process
E(NH3) = -148492.43 kJ/mol
2*E(NH3) = -296984.86 kJ/mol
E(N2) = -287555.62 kJ/mol
E(H2) = -3094.26 kJ/mol
3*E(H2) = -9282.78 kJ/mol
ΔE = 2*E(NH3) - [E(N2) + 3*E(H2)] = -146.46 kJ/mol
The Haber Process is an exothermic reaction. However, the literature value is quoted to be -45.940 kJ/mol so our value is greatly deviated. [1]
- ↑ Klemola, K., Chemical Reaction Equilibrium Calculation Task For ChE Undergraduates–Simulating Fritz Haber’s Ammonia Synthesis With Thermodynamic Software. Chemical Engineering Education, 48(2), pp.115-120
HCl
test molecule |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d.p) |
| Final Energy | -460.80077875 a.u. |
| RMS Gradient Norm | 0.00005211 a.u. |
| Point Group | C∞V |
| Bond Length | 1.10550 Å |
Charge distribution on H = + 0.284
Charge distribution on Cl = - 0.284
Chlorine atom is more electronegative than Hydrogen atom thus has a higher tendency of attracting a pair of bonding electrons.
Optimisation Table
Item Value Threshold Converged?
Maximum Force 0.000090 0.000450 YES
RMS Force 0.000090 0.000300 YES
Maximum Displacement 0.000139 0.001800 YES
RMS Displacement 0.000197 0.001200 YES
Predicted change in Energy=-1.256951D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.286 -DE/DX = 0.0001 !
--------------------------------------------------------------------------------
Vibrational Modes of HCl

HCl is a linear molecule, using the rule 3N-5 where N = 2, we would expect it to only have 1 vibrational mode.
IR Spectrum of HCl
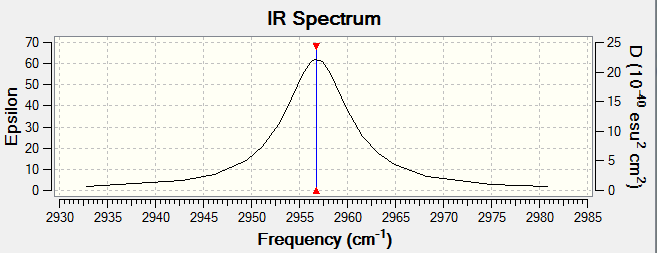
HCl has a change in dipole moment therefore it is IR active. It only has one peak as predicted and it is a stretching frequency at around 2957 cm-1.
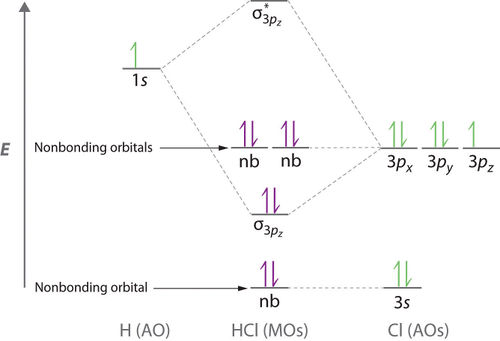
Molecular Orbital Analysis of HCl
The Cl electrons residing up to 3s orbitals (1s, 2s, 2px, 2py, 2pz, 3s) are largely stabilised than the electron in 1s orbital of H atom, therefore they cannot mix and form bond. The 3p electron in Cl atom has similar energy to 1s electron in H atom therefore they can mix. However, due to the different symmetries of 3px and 3py orbitals compared to that of 1s orbital, the only possible mixing is σ overlap between 1s orbital of H atom and 3pz orbital of Cl atom. Both of these orbitals are half-filled thus are allowed to form a bond.
The bond order of HCl is 1 because there are 2 electrons in the bonding orbital but none in the non-bonding orbital.
Molecular Coefficient Table of HCl
1 2 3 4 5
O O O O O
Eigenvalues -- -101.55786 -9.47437 -7.23847 -7.22836 -7.22836
1 1 Cl 1S 0.99600 -0.28467 -0.00189 0.00000 0.00000
2 2S 0.01517 1.02220 0.00687 0.00000 0.00000
3 2PX 0.00000 0.00000 0.00000 0.99117 0.00000
4 2PY 0.00000 0.00000 0.00000 0.00000 0.99117
5 2PZ -0.00004 -0.00630 0.99047 0.00000 0.00000
6 3S -0.02109 0.07379 0.00034 0.00000 0.00000
7 3PX 0.00000 0.00000 0.00000 0.02799 0.00000
8 3PY 0.00000 0.00000 0.00000 0.00000 0.02799
9 3PZ -0.00002 -0.00120 0.03045 0.00000 0.00000
10 4S 0.00171 -0.01358 0.00215 0.00000 0.00000
11 4PX 0.00000 0.00000 0.00000 -0.00736 0.00000
12 4PY 0.00000 0.00000 0.00000 0.00000 -0.00736
13 4PZ -0.00010 0.00178 -0.00945 0.00000 0.00000
14 5XX 0.00757 -0.01669 0.00002 0.00000 0.00000
15 5YY 0.00757 -0.01669 0.00002 0.00000 0.00000
16 5ZZ 0.00765 -0.01573 -0.00228 0.00000 0.00000
17 5XY 0.00000 0.00000 0.00000 0.00000 0.00000
18 5XZ 0.00000 0.00000 0.00000 -0.00048 0.00000
19 5YZ 0.00000 0.00000 0.00000 0.00000 -0.00048
20 2 H 1S 0.00005 -0.00124 0.00256 0.00000 0.00000
21 2S -0.00022 0.00300 -0.00359 0.00000 0.00000
22 3PX 0.00000 0.00000 0.00000 -0.00039 0.00000
23 3PY 0.00000 0.00000 0.00000 0.00000 -0.00039
24 3PZ -0.00014 -0.00052 0.00180 0.00000 0.00000
6 7 8 9 10
O O O O V
Eigenvalues -- -0.84773 -0.47433 -0.33163 -0.33163 0.01366
1 1 Cl 1S 0.08094 0.02733 0.00000 0.00000 -0.02450
2 2S -0.36382 -0.12619 0.00000 0.00000 0.10936
3 2PX 0.00000 0.00000 -0.28537 0.00000 0.00000
4 2PY 0.00000 0.00000 0.00000 -0.28537 0.00000
5 2PZ 0.04545 -0.23220 0.00000 0.00000 -0.16897
6 3S 0.73327 0.26260 0.00000 0.00000 -0.26941
7 3PX 0.00000 0.00000 0.73004 0.00000 0.00000
8 3PY 0.00000 0.00000 0.00000 0.73004 0.00000
9 3PZ -0.10755 0.57863 0.00000 0.00000 0.46375
10 4S 0.27021 0.24735 0.00000 0.00000 -0.72409
11 4PX 0.00000 0.00000 0.40430 0.00000 0.00000
12 4PY 0.00000 0.00000 0.00000 0.40430 0.00000
13 4PZ -0.01216 0.16969 0.00000 0.00000 0.87011
14 5XX -0.02029 0.01824 0.00000 0.00000 -0.04398
15 5YY -0.02029 0.01824 0.00000 0.00000 -0.04398
16 5ZZ 0.02127 -0.05761 0.00000 0.00000 0.10073
17 5XY 0.00000 0.00000 0.00000 0.00000 0.00000
18 5XZ 0.00000 0.00000 -0.01866 0.00000 0.00000
19 5YZ 0.00000 0.00000 0.00000 -0.01866 0.00000
20 2 H 1S 0.15662 -0.28655 0.00000 0.00000 0.24914
21 2S 0.04977 -0.24977 0.00000 0.00000 1.50879
22 3PX 0.00000 0.00000 0.01771 0.00000 0.00000
23 3PY 0.00000 0.00000 0.00000 0.01771 0.00000
24 3PZ 0.01666 -0.01719 0.00000 0.00000 -0.01235
11 12 13 14 15
V V V V V
Eigenvalues -- 0.36800 0.40255 0.43685 0.43685 0.68586
1 1 Cl 1S -0.05961 -0.02916 0.00000 0.00000 0.03175
2 2S 0.04945 0.06099 0.00000 0.00000 -0.03265
3 2PX 0.00000 0.00000 0.30181 0.00000 0.00000
4 2PY 0.00000 0.00000 0.00000 0.30181 0.00000
5 2PZ -0.01321 0.23797 0.00000 0.00000 0.18170
6 3S -1.26542 -0.52542 0.00000 0.00000 0.69879
7 3PX 0.00000 0.00000 -1.16096 0.00000 0.00000
8 3PY 0.00000 0.00000 0.00000 -1.16096 0.00000
9 3PZ 0.11244 -0.94335 0.00000 0.00000 -0.68364
10 4S 1.96653 0.54265 0.00000 0.00000 -1.77015
11 4PX 0.00000 0.00000 1.23592 0.00000 0.00000
12 4PY 0.00000 0.00000 0.00000 1.23592 0.00000
13 4PZ -0.65815 1.18628 0.00000 0.00000 1.39906
14 5XX 0.01785 -0.10153 0.00000 0.00000 0.22144
15 5YY 0.01785 -0.10153 0.00000 0.00000 0.22144
16 5ZZ -0.34080 0.11877 0.00000 0.00000 -0.36870
17 5XY 0.00000 0.00000 0.00000 0.00000 0.00000
18 5XZ 0.00000 0.00000 0.01458 0.00000 0.00000
19 5YZ 0.00000 0.00000 0.00000 0.01458 0.00000
20 2 H 1S -0.37113 0.25587 0.00000 0.00000 -0.86140
21 2S -0.41553 -0.05089 0.00000 0.00000 2.18749
22 3PX 0.00000 0.00000 -0.00153 0.00000 0.00000
23 3PY 0.00000 0.00000 0.00000 -0.00153 0.00000
24 3PZ -0.04561 0.04911 0.00000 0.00000 0.01578
16 17 18 19 20
V V V V V
Eigenvalues -- 0.80005 0.80005 0.86468 0.86468 1.16034
1 1 Cl 1S 0.00000 0.00000 0.00000 0.00000 0.00066
2 2S 0.00000 0.00000 0.00000 0.00000 0.03128
3 2PX 0.00000 -0.01332 0.00000 0.00000 0.00000
4 2PY -0.01332 0.00000 0.00000 0.00000 0.00000
5 2PZ 0.00000 0.00000 0.00000 0.00000 -0.01614
6 3S 0.00000 0.00000 0.00000 0.00000 0.07339
7 3PX 0.00000 0.03860 0.00000 0.00000 0.00000
8 3PY 0.03860 0.00000 0.00000 0.00000 0.00000
9 3PZ 0.00000 0.00000 0.00000 0.00000 -0.00125
10 4S 0.00000 0.00000 0.00000 0.00000 -0.17360
11 4PX 0.00000 0.01600 0.00000 0.00000 0.00000
12 4PY 0.01600 0.00000 0.00000 0.00000 0.00000
13 4PZ 0.00000 0.00000 0.00000 0.00000 0.24726
14 5XX 0.00000 0.00000 0.86603 0.00000 0.46825
15 5YY 0.00000 0.00000 -0.86603 0.00000 0.46825
16 5ZZ 0.00000 0.00000 0.00000 0.00000 -0.82390
17 5XY 0.00000 0.00000 0.00000 1.00000 0.00000
18 5XZ 0.00000 0.95574 0.00000 0.00000 0.00000
19 5YZ 0.95574 0.00000 0.00000 0.00000 0.00000
20 2 H 1S 0.00000 0.00000 0.00000 0.00000 0.92285
21 2S 0.00000 0.00000 0.00000 0.00000 -0.40892
22 3PX 0.00000 -0.17261 0.00000 0.00000 0.00000
23 3PY -0.17261 0.00000 0.00000 0.00000 0.00000
24 3PZ 0.00000 0.00000 0.00000 0.00000 -0.15003
21 22 23 24
V V V V
Eigenvalues -- 2.05856 2.05856 2.75279 4.27800
1 1 Cl 1S 0.00000 0.00000 -0.02965 0.16933
2 2S 0.00000 0.00000 0.15175 -0.79433
3 2PX 0.00000 0.01699 0.00000 0.00000
4 2PY 0.01699 0.00000 0.00000 0.00000
5 2PZ 0.00000 0.00000 -0.14494 -0.03702
6 3S 0.00000 0.00000 -0.56866 5.51631
7 3PX 0.00000 -0.02940 0.00000 0.00000
8 3PY -0.02940 0.00000 0.00000 0.00000
9 3PZ 0.00000 0.00000 0.58914 0.15492
10 4S 0.00000 0.00000 -0.52879 0.10829
11 4PX 0.00000 -0.15059 0.00000 0.00000
12 4PY -0.15059 0.00000 0.00000 0.00000
13 4PZ 0.00000 0.00000 0.26345 -0.05395
14 5XX 0.00000 0.00000 0.47810 -2.41449
15 5YY 0.00000 0.00000 0.47810 -2.41449
16 5ZZ 0.00000 0.00000 -0.47857 -2.60306
17 5XY 0.00000 0.00000 0.00000 0.00000
18 5XZ 0.00000 0.34228 0.00000 0.00000
19 5YZ 0.34228 0.00000 0.00000 0.00000
20 2 H 1S 0.00000 0.00000 0.27323 0.13310
21 2S 0.00000 0.00000 0.64729 -0.07000
22 3PX 0.00000 1.01311 0.00000 0.00000
23 3PY 1.01311 0.00000 0.00000 0.00000
24 3PZ 0.00000 0.00000 1.23461 0.27943
Density Matrix:
1 2 3 4 5
1 1 Cl 1S 2.16070
2 2S -0.61758 2.38693
3 2PX 0.00000 0.00000 2.12770
4 2PY 0.00000 0.00000 0.00000 2.12770
5 2PZ -0.00558 0.02626 0.00000 0.00000 2.07410
6 3S 0.04903 -0.44960 0.00000 0.00000 -0.05556
7 3PX 0.00000 0.00000 -0.36119 0.00000 0.00000
8 3PY 0.00000 0.00000 0.00000 -0.36119 0.00000
9 3PZ 0.01475 -0.06982 0.00000 0.00000 -0.21815
10 4S 0.06840 -0.28673 0.00000 0.00000 -0.08588
11 4PX 0.00000 0.00000 -0.24535 0.00000 0.00000
12 4PY 0.00000 0.00000 0.00000 -0.24535 0.00000
13 4PZ 0.00613 -0.03048 0.00000 0.00000 -0.09866
14 5XX 0.02229 -0.02373 0.00000 0.00000 -0.01006
15 5YY 0.02229 -0.02373 0.00000 0.00000 -0.01006
16 5ZZ 0.02449 -0.03288 0.00000 0.00000 0.02438
17 5XY 0.00000 0.00000 0.00000 0.00000 0.00000
18 5XZ 0.00000 0.00000 0.00970 0.00000 0.00000
19 5YZ 0.00000 0.00000 0.00000 0.00970 0.00000
20 2 H 1S 0.01049 -0.04413 0.00000 0.00000 0.15240
21 2S -0.00772 0.03290 0.00000 0.00000 0.11337
22 3PX 0.00000 0.00000 -0.01088 0.00000 0.00000
23 3PY 0.00000 0.00000 0.00000 -0.01088 0.00000
24 3PZ 0.00176 -0.00884 0.00000 0.00000 0.01307
6 7 8 9 10
6 3S 1.22508
7 3PX 0.00000 1.06750
8 3PY 0.00000 0.00000 1.06750
9 3PZ 0.14602 0.00000 0.00000 0.69461
10 4S 0.52411 0.00000 0.00000 0.22828 0.26877
11 4PX 0.00000 0.58991 0.00000 0.00000 0.00000
12 4PY 0.00000 0.00000 0.58991 0.00000 0.00000
13 4PZ 0.07155 0.00000 0.00000 0.19841 0.07728
14 5XX -0.02296 0.00000 0.00000 0.02551 -0.00146
15 5YY -0.02296 0.00000 0.00000 0.02551 -0.00146
16 5ZZ -0.00172 0.00000 0.00000 -0.07134 -0.01656
17 5XY 0.00000 0.00000 0.00000 0.00000 0.00000
18 5XZ 0.00000 -0.02727 0.00000 0.00000 0.00000
19 5YZ 0.00000 0.00000 -0.02727 0.00000 0.00000
20 2 H 1S 0.07901 0.00000 0.00000 -0.36515 -0.05707
21 2S -0.05774 0.00000 0.00000 -0.29997 -0.09676
22 3PX 0.00000 0.02584 0.00000 0.00000 0.00000
23 3PY 0.00000 0.00000 0.02584 0.00000 0.00000
24 3PZ 0.01534 0.00000 0.00000 -0.02336 0.00052
11 12 13 14 15
11 4PX 0.32703
12 4PY 0.00000 0.32703
13 4PZ 0.00000 0.00000 0.05807
14 5XX 0.00000 0.00000 0.00662 0.00216
15 5YY 0.00000 0.00000 0.00662 0.00216 0.00216
16 5ZZ 0.00000 0.00000 -0.02008 -0.00232 -0.00232
17 5XY 0.00000 0.00000 0.00000 0.00000 0.00000
18 5XZ -0.01508 0.00000 0.00000 0.00000 0.00000
19 5YZ 0.00000 -0.01508 0.00000 0.00000 0.00000
20 2 H 1S 0.00000 0.00000 -0.10111 -0.01677 -0.01677
21 2S 0.00000 0.00000 -0.08590 -0.01123 -0.01123
22 3PX 0.01433 0.00000 0.00000 0.00000 0.00000
23 3PY 0.00000 0.01433 0.00000 0.00000 0.00000
24 3PZ 0.00000 0.00000 -0.00627 -0.00129 -0.00129
16 17 18 19 20
16 5ZZ 0.00816
17 5XY 0.00000 0.00000
18 5XZ 0.00000 0.00000 0.00070
19 5YZ 0.00000 0.00000 0.00000 0.00070
20 2 H 1S 0.03971 0.00000 0.00000 0.00000 0.21331
21 2S 0.03081 0.00000 0.00000 0.00000 0.15871
22 3PX 0.00000 0.00000 -0.00066 0.00000 0.00000
23 3PY 0.00000 0.00000 0.00000 -0.00066 0.00000
24 3PZ 0.00270 0.00000 0.00000 0.00000 0.01508
21 22 23 24
21 2S 0.12976
22 3PX 0.00000 0.00063
23 3PY 0.00000 0.00000 0.00063
24 3PZ 0.01023 0.00000 0.00000 0.00115
CH3OH
test molecule |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d.p) |
| Final Energy | -115.72396437 a.u. |
| RMS Gradient Norm | 0.00001494 a.u. |
| Point Group | C1 |
Optimisation Table
Item Value Threshold Converged?
Maximum Force 0.000038 0.000450 YES
RMS Force 0.000020 0.000300 YES
Maximum Displacement 0.000497 0.001800 YES
RMS Displacement 0.000240 0.001200 YES
Predicted change in Energy=-1.610243D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1006 -DE/DX = 0.0 !
! R2 R(1,3) 1.1006 -DE/DX = 0.0 !
! R3 R(1,4) 1.093 -DE/DX = 0.0 !
! R4 R(1,5) 1.4181 -DE/DX = 0.0 !
! R5 R(5,6) 0.9652 -DE/DX = 0.0 !
! A1 A(2,1,3) 108.2583 -DE/DX = 0.0 !
! A2 A(2,1,4) 107.8992 -DE/DX = 0.0 !
! A3 A(2,1,5) 112.8281 -DE/DX = 0.0 !
! A4 A(3,1,4) 107.8997 -DE/DX = 0.0 !
! A5 A(3,1,5) 112.8274 -DE/DX = 0.0 !
! A6 A(4,1,5) 106.9041 -DE/DX = 0.0 !
! A7 A(1,5,6) 107.7375 -DE/DX = 0.0 !
! D1 D(2,1,5,6) 61.5104 -DE/DX = 0.0 !
! D2 D(3,1,5,6) -61.5824 -DE/DX = 0.0 !
! D3 D(4,1,5,6) 179.9638 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Vibrational Modes of CH3OH
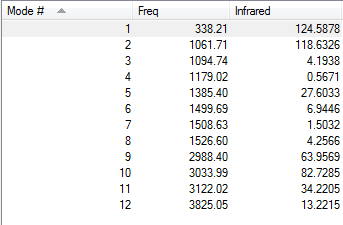
CH3OH is a non-linear molecule, using the rule 3N-6 where N = 6, we would expect it to have 12 vibrational modes.
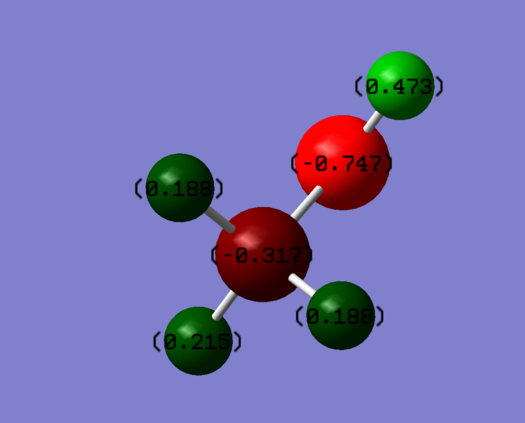
The oxygen atom is more electronegative than carbon and hydrogen. Therefore, it is most negatively charged.
IR Spectrum of CH3OH
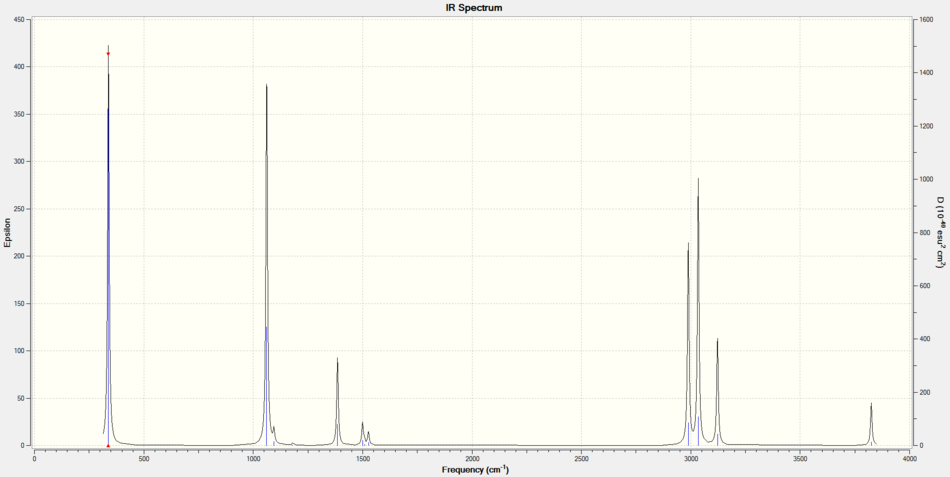
Molecular Orbital Analysis of CH3OH
Molecular Coefficient Table of CH3OH
1 2 3 4 5
O O O O O
Eigenvalues -- -19.13323 -10.21741 -1.00348 -0.68461 -0.49746
1 1 C 1S 0.00001 0.99288 -0.07451 -0.18213 0.02296
2 2S 0.00021 0.04924 0.14082 0.36746 -0.04378
3 2PX -0.00033 -0.00049 -0.10035 0.06600 -0.22267
4 2PY 0.00010 0.00001 0.00321 0.00812 0.22340
5 2PZ 0.00000 0.00000 0.00000 -0.00001 0.00006
6 3S -0.00162 -0.01419 0.04858 0.31204 -0.04524
7 3PX 0.00074 -0.00029 0.00250 0.02863 -0.07688
8 3PY -0.00050 0.00010 -0.00814 -0.00520 0.09766
9 3PZ 0.00000 0.00000 0.00001 0.00001 -0.00001
10 4XX 0.00032 -0.00902 0.01802 -0.01507 0.02270
11 4YY 0.00006 -0.00907 -0.01070 -0.00605 -0.00255
12 4ZZ 0.00010 -0.00910 -0.01132 -0.00767 -0.01465
13 4XY -0.00006 0.00003 -0.00064 -0.00132 -0.00965
14 4XZ 0.00000 0.00000 0.00000 0.00000 0.00000
15 4YZ 0.00000 0.00000 0.00000 0.00000 -0.00004
16 2 H 1S 0.00000 -0.00017 0.02933 0.13947 -0.11858
17 2S -0.00004 0.00278 0.00343 0.04344 -0.07603
18 3PX 0.00005 0.00004 -0.00227 -0.00251 -0.00111
19 3PY -0.00001 -0.00004 0.00252 0.00644 0.00032
20 3PZ -0.00006 -0.00005 0.00361 0.01031 -0.00567
21 3 H 1S 0.00000 -0.00017 0.02933 0.13947 -0.11817
22 2S -0.00004 0.00278 0.00342 0.04343 -0.07571
23 3PX 0.00005 0.00004 -0.00227 -0.00251 -0.00112
24 3PY -0.00001 -0.00004 0.00251 0.00640 0.00035
25 3PZ 0.00006 0.00005 -0.00362 -0.01034 0.00567
26 4 H 1S 0.00012 -0.00020 0.02757 0.14807 0.03097
27 2S 0.00024 0.00271 0.00894 0.05050 0.01704
28 3PX 0.00005 0.00005 -0.00324 -0.00427 -0.00428
29 3PY 0.00003 0.00008 -0.00363 -0.01207 0.00174
30 3PZ 0.00000 0.00000 0.00001 0.00002 0.00001
31 5 O 1S 0.99282 -0.00011 -0.20324 0.06132 -0.04071
32 2S 0.02609 -0.00019 0.44780 -0.14040 0.09438
33 2PX 0.00039 -0.00010 0.05685 0.17576 0.28950
34 2PY -0.00105 0.00002 -0.09528 0.08703 0.31404
35 2PZ 0.00000 0.00000 0.00000 0.00000 0.00002
36 3S 0.01107 0.00199 0.42747 -0.16902 0.15384
37 3PX 0.00020 0.00021 0.03906 0.07928 0.13067
38 3PY -0.00005 0.00024 -0.03356 0.03331 0.16062
39 3PZ 0.00000 0.00000 0.00000 0.00000 0.00002
40 4XX -0.00796 -0.00076 0.00845 0.01600 0.01182
41 4YY -0.00771 -0.00021 -0.00046 -0.00655 -0.02095
42 4ZZ -0.00787 -0.00004 -0.01011 0.00059 0.00125
43 4XY 0.00011 0.00006 -0.00224 -0.00712 -0.00043
44 4XZ 0.00000 0.00000 0.00000 0.00000 0.00000
45 4YZ 0.00000 0.00000 0.00000 0.00000 0.00000
46 6 H 1S 0.00020 0.00009 0.14498 -0.12336 -0.21334
47 2S -0.00125 0.00042 0.01716 -0.04421 -0.13834
48 3PX -0.00017 0.00019 0.01174 -0.00279 -0.00242
49 3PY -0.00024 0.00006 0.01995 -0.01282 -0.01181
50 3PZ 0.00000 0.00000 0.00000 0.00000 0.00000
6 7 8 9 10
O O O O V
Eigenvalues -- -0.43915 -0.42485 -0.32471 -0.26079 0.07798
1 1 C 1S -0.00001 0.00131 0.01359 0.00000 0.02959
2 2S 0.00002 -0.00187 -0.03365 -0.00001 -0.02512
3 2PX 0.00009 0.27083 0.13524 0.00000 -0.20836
4 2PY 0.00004 0.32478 -0.24525 0.00000 0.07382
5 2PZ 0.42861 -0.00009 0.00001 -0.17477 0.00018
6 3S 0.00004 -0.01338 -0.03450 0.00000 -0.64815
7 3PX 0.00004 0.11307 0.05295 -0.00001 -0.56875
8 3PY -0.00003 0.15200 -0.05835 -0.00003 0.15211
9 3PZ 0.16711 -0.00008 -0.00002 -0.02347 0.00059
10 4XX -0.00001 -0.01627 -0.01219 0.00000 -0.00929
11 4YY -0.00008 0.01874 -0.00186 0.00005 0.01739
12 4ZZ 0.00008 -0.00320 0.01489 -0.00005 0.01214
13 4XY -0.00001 0.01178 -0.02354 0.00001 -0.00307
14 4XZ 0.00232 -0.00001 0.00001 -0.03018 0.00000
15 4YZ -0.01791 -0.00008 0.00007 0.01376 0.00001
16 2 H 1S -0.20969 -0.05254 0.10939 0.14039 0.01328
17 2S -0.17486 -0.03688 0.13390 0.18081 0.49819
18 3PX 0.00402 0.00494 0.00140 0.00124 -0.00640
19 3PY -0.00583 0.00435 -0.00314 0.00298 -0.00299
20 3PZ -0.00200 -0.00245 0.00519 0.00096 -0.00311
21 3 H 1S 0.21016 -0.05194 0.10881 -0.14070 0.01317
22 2S 0.17518 -0.03635 0.13328 -0.18125 0.49651
23 3PX -0.00402 0.00493 0.00140 -0.00124 -0.00639
24 3PY 0.00580 0.00438 -0.00317 -0.00296 -0.00296
25 3PZ -0.00205 0.00243 -0.00518 0.00098 0.00313
26 4 H 1S -0.00041 0.25402 -0.15184 0.00031 0.00083
27 2S -0.00030 0.19559 -0.16657 0.00044 0.26070
28 3PX 0.00001 -0.00164 0.00369 0.00000 -0.00791
29 3PY 0.00002 -0.00657 0.00189 -0.00002 0.00551
30 3PZ 0.00821 0.00002 -0.00001 -0.00488 0.00000
31 5 O 1S 0.00000 -0.02434 -0.06721 0.00000 0.09248
32 2S 0.00000 0.04443 0.12908 0.00000 -0.11726
33 2PX -0.00010 -0.31801 -0.23119 0.00000 -0.00118
34 2PY -0.00004 0.08938 0.43763 0.00002 0.24154
35 2PZ 0.23661 -0.00005 0.00000 0.60340 -0.00005
36 3S 0.00000 0.10860 0.33329 -0.00001 -1.13040
37 3PX -0.00005 -0.17683 -0.16355 0.00000 -0.00479
38 3PY -0.00001 0.05959 0.29444 0.00002 0.43222
39 3PZ 0.14916 -0.00003 0.00000 0.46617 -0.00012
40 4XX 0.00000 -0.00831 -0.00940 0.00000 0.03762
41 4YY 0.00000 -0.00332 -0.03165 0.00000 0.02713
42 4ZZ 0.00000 -0.00106 -0.00037 0.00000 0.04823
43 4XY 0.00000 0.02248 0.00949 0.00000 0.00576
44 4XZ 0.01366 0.00000 0.00000 0.00785 0.00000
45 4YZ -0.01065 0.00000 0.00001 -0.02087 0.00001
46 6 H 1S 0.00004 0.06301 -0.14490 0.00000 0.10846
47 2S 0.00003 0.05598 -0.11413 0.00001 1.20083
48 3PX 0.00000 -0.00888 -0.01139 0.00000 -0.00180
49 3PY 0.00001 0.00797 0.00357 0.00000 -0.00626
50 3PZ 0.00785 0.00000 0.00000 0.02027 0.00000
11 12 13 14 15
V V V V V
Eigenvalues -- 0.13723 0.17800 0.18061 0.20050 0.53040
1 1 C 1S -0.15536 -0.02280 0.00002 0.05326 -0.03207
2 2S 0.21015 0.01815 0.00008 -0.08368 0.35618
3 2PX 0.05651 -0.12807 0.00019 0.29837 -0.94756
4 2PY 0.02528 -0.39652 0.00270 -0.11638 0.15128
5 2PZ 0.00009 -0.00275 -0.44040 -0.00086 -0.00018
6 3S 2.53039 0.31720 -0.00063 -0.57274 -0.42536
7 3PX 0.15339 -0.53141 0.00105 1.18175 1.46385
8 3PY 0.07860 -1.28555 0.00904 -0.54116 -0.26872
9 3PZ 0.00034 -0.00844 -1.37065 -0.00240 0.00039
10 4XX -0.01761 -0.01022 0.00001 0.02795 -0.04932
11 4YY -0.00829 0.01863 -0.00018 -0.00942 0.03103
12 4ZZ -0.01053 -0.01057 0.00020 -0.02600 0.01608
13 4XY -0.00238 0.01032 -0.00006 -0.00905 0.01230
14 4XZ 0.00000 -0.00001 0.00046 0.00000 0.00000
15 4YZ 0.00001 -0.00020 -0.01633 -0.00007 -0.00003
16 2 H 1S -0.01127 -0.03350 -0.06658 -0.06927 -0.11590
17 2S -1.06208 -0.90464 -1.59331 -0.41096 -0.10462
18 3PX -0.00227 -0.00557 -0.00071 0.01213 -0.01353
19 3PY 0.00187 -0.00810 0.00798 -0.00302 -0.00862
20 3PZ 0.00357 0.00557 -0.00081 0.00707 -0.00988
21 3 H 1S -0.01137 -0.03241 0.06738 -0.06897 -0.11594
22 2S -1.06318 -0.87889 1.60929 -0.40333 -0.10464
23 3PX -0.00227 -0.00556 0.00074 0.01214 -0.01351
24 3PY 0.00186 -0.00825 -0.00783 -0.00308 -0.00859
25 3PZ -0.00358 -0.00555 -0.00066 -0.00707 0.00993
26 4 H 1S -0.02806 0.10273 -0.00078 -0.01823 -0.04607
27 2S -1.25438 1.77073 -0.01542 0.35408 -0.06304
28 3PX -0.00405 -0.00048 -0.00003 0.01090 -0.00906
29 3PY -0.00338 0.00508 -0.00006 -0.00594 0.01155
30 3PZ 0.00001 -0.00012 -0.01360 -0.00002 -0.00002
31 5 O 1S 0.05194 0.01633 0.00003 -0.03893 0.00749
32 2S -0.03882 -0.01689 -0.00011 0.05855 -0.18076
33 2PX -0.08207 -0.12761 0.00013 0.38734 -0.27444
34 2PY 0.11098 0.05772 -0.00049 0.15189 0.07813
35 2PZ -0.00003 0.00049 0.08465 0.00013 -0.00004
36 3S -0.73062 -0.26711 -0.00001 0.49192 0.83388
37 3PX -0.21740 -0.23975 0.00001 0.82116 0.24758
38 3PY 0.18231 0.16989 -0.00133 0.30567 -0.01733
39 3PZ -0.00006 0.00132 0.21561 0.00036 -0.00005
40 4XX 0.03692 0.00227 0.00000 0.00181 0.10533
41 4YY 0.02495 0.00713 -0.00004 -0.01929 -0.08947
42 4ZZ 0.02908 0.00481 0.00006 -0.01450 -0.09025
43 4XY 0.00836 -0.00961 0.00008 -0.01299 -0.00919
44 4XZ 0.00000 -0.00006 -0.00930 -0.00002 0.00000
45 4YZ 0.00001 -0.00013 -0.01266 -0.00004 0.00000
46 6 H 1S 0.06015 -0.02915 0.00008 0.06030 -0.05108
47 2S 0.49339 -0.14472 0.00025 0.96821 0.23025
48 3PX -0.00931 0.00508 -0.00007 0.00805 -0.03974
49 3PY -0.00466 0.00014 0.00002 -0.00133 0.01760
50 3PZ 0.00000 0.00007 0.00728 0.00002 0.00000
16 17 18 19 20
V V V V V
Eigenvalues -- 0.55483 0.56057 0.77259 0.84988 0.85234
1 1 C 1S -0.00232 0.00001 0.00732 0.00017 -0.03574
2 2S 0.03318 -0.00007 -0.00717 0.00032 -0.01598
3 2PX -0.17397 0.00020 -0.00289 -0.00042 0.08669
4 2PY -0.69411 -0.00005 0.39930 -0.00380 0.58601
5 2PZ 0.00004 -0.70643 -0.00047 -0.75949 -0.00437
6 3S -0.05118 0.00002 -0.17302 -0.00224 0.32720
7 3PX 0.39644 -0.00046 -0.13127 0.00040 -0.06901
8 3PY 1.68561 0.00016 -0.90711 0.00805 -1.28304
9 3PZ -0.00017 1.76292 0.00075 1.51059 0.00870
10 4XX 0.00607 -0.00001 -0.09067 -0.00009 0.01752
11 4YY -0.06564 0.00041 0.00323 -0.00005 -0.14296
12 4ZZ 0.06596 -0.00040 0.07802 0.00041 0.06846
13 4XY -0.03698 0.00005 -0.13019 0.00080 -0.13723
14 4XZ 0.00005 -0.00408 0.00013 0.14429 0.00092
15 4YZ 0.00046 0.08430 0.00024 -0.19800 -0.00039
16 2 H 1S 0.20807 0.39723 0.27912 -0.63935 0.04535
17 2S 0.21622 0.45275 -0.45390 1.53314 -0.67010
18 3PX -0.00137 -0.00878 0.01548 -0.05536 0.03637
19 3PY 0.03439 -0.00022 -0.01593 0.06166 0.00825
20 3PZ 0.00083 0.03553 -0.03362 0.05974 -0.01558
21 3 H 1S 0.20660 -0.39792 0.27896 0.63933 0.05091
22 2S 0.21483 -0.45362 -0.45253 -1.52747 -0.68312
23 3PX -0.00133 0.00881 0.01543 0.05508 0.03681
24 3PY 0.03437 0.00022 -0.01581 -0.06149 0.00777
25 3PZ -0.00083 0.03553 0.03362 0.05985 0.01604
26 4 H 1S -0.48808 0.00087 -0.21430 0.00326 -0.78448
27 2S -0.48004 0.00105 0.93961 -0.00551 1.43449
28 3PX 0.01406 -0.00003 -0.05080 0.00027 -0.07292
29 3PY 0.03249 0.00001 -0.02530 0.00030 -0.08845
30 3PZ 0.00000 0.03737 0.00002 -0.03583 0.00002
31 5 O 1S 0.01221 0.00000 0.02194 0.00000 -0.00162
32 2S -0.14822 0.00019 0.00642 0.00141 -0.22225
33 2PX 0.07693 -0.00005 -0.09040 -0.00040 0.02976
34 2PY -0.04839 -0.00013 0.36634 0.00255 -0.51566
35 2PZ 0.00004 -0.19458 0.00012 0.25655 0.00142
36 3S 0.19995 -0.00038 0.06568 -0.00162 0.22451
37 3PX -0.00596 -0.00002 0.25717 0.00068 -0.07718
38 3PY -0.20266 0.00007 -0.08122 -0.00356 0.70508
39 3PZ 0.00001 -0.19024 -0.00016 -0.36206 -0.00205
40 4XX -0.03136 0.00003 0.04149 0.00048 -0.07924
41 4YY -0.02154 0.00014 0.20064 0.00096 -0.17194
42 4ZZ -0.04417 -0.00001 -0.05970 0.00033 -0.05572
43 4XY 0.01822 0.00000 0.18127 -0.00010 -0.01133
44 4XZ 0.00001 -0.00752 -0.00006 -0.07956 -0.00048
45 4YZ 0.00009 0.02130 -0.00005 0.02313 0.00005
46 6 H 1S 0.12975 0.00007 0.82473 0.00041 -0.11306
47 2S 0.11707 -0.00030 -0.81012 -0.00098 0.24625
48 3PX -0.04625 0.00004 -0.06194 -0.00027 0.06852
49 3PY -0.00811 0.00000 -0.14721 -0.00046 0.09888
50 3PZ -0.00002 -0.00166 0.00004 0.00133 0.00008
21 22 23 24 25
V V V V V
Eigenvalues -- 0.90348 0.94617 0.95971 1.01879 1.07945
1 1 C 1S -0.09346 -0.00719 0.00002 -0.06226 -0.02722
2 2S 0.39581 -0.90290 0.00086 -1.56578 -0.54258
3 2PX 0.27957 -0.26995 0.00022 -0.10628 0.02063
4 2PY -0.19470 -0.08323 0.00009 -0.02937 0.26605
5 2PZ 0.00020 -0.00016 -0.08658 0.00011 -0.00048
6 3S -0.68667 2.41448 -0.00222 3.61877 1.64989
7 3PX -0.13950 0.27177 -0.00034 0.78389 -0.21965
8 3PY 0.22888 -0.15362 0.00028 0.09142 -0.13232
9 3PZ -0.00043 0.00000 -0.30520 -0.00044 0.00173
10 4XX 0.00421 0.01945 0.00003 -0.21186 0.08252
11 4YY -0.02224 0.08928 -0.00080 -0.06033 -0.23383
12 4ZZ -0.15535 -0.13870 0.00085 0.00201 0.11698
13 4XY 0.02008 0.00139 -0.00007 0.04395 0.09749
14 4XZ -0.00012 -0.00010 -0.01827 0.00002 0.00011
15 4YZ -0.00038 -0.00086 -0.13805 0.00019 0.00096
16 2 H 1S -0.72504 -0.06962 -0.23037 -0.02975 -0.03483
17 2S 0.73990 -0.47865 0.17011 -1.01483 -0.41198
18 3PX -0.01215 0.02247 0.04897 -0.00980 -0.03046
19 3PY 0.03116 -0.05390 0.03902 0.00257 0.06038
20 3PZ 0.09141 0.01116 0.00329 -0.03399 -0.06254
21 3 H 1S -0.72463 -0.06820 0.23116 -0.02991 -0.03529
22 2S 0.73931 -0.48042 -0.16954 -1.01440 -0.41335
23 3PX -0.01225 0.02226 -0.04909 -0.00976 -0.03017
24 3PY 0.03074 -0.05416 -0.03870 0.00274 0.06078
25 3PZ -0.09151 -0.01092 0.00369 0.03398 0.06232
26 4 H 1S -0.36725 0.37692 -0.00083 -0.10035 -0.35029
27 2S 0.09999 -0.95562 0.00121 -1.09206 0.16450
28 3PX -0.01131 0.00015 0.00010 0.00250 0.09085
29 3PY -0.05504 0.05855 -0.00026 0.02825 -0.06929
30 3PZ 0.00006 -0.00030 -0.07179 -0.00002 0.00032
31 5 O 1S 0.00383 0.00484 -0.00001 -0.00879 -0.01875
32 2S -0.44532 -0.72044 0.00056 0.01684 -0.49979
33 2PX -0.24163 0.41522 -0.00015 -0.55588 0.33629
34 2PY 0.12664 -0.24505 0.00038 0.02540 0.54218
35 2PZ -0.00021 -0.00080 -0.89330 -0.00016 0.00036
36 3S 0.84339 1.12566 -0.00078 -0.28445 1.04736
37 3PX 0.27739 -0.67856 0.00021 0.76800 -1.00151
38 3PY -0.11231 0.39630 -0.00073 0.03929 -1.21027
39 3PZ 0.00026 0.00094 1.10743 0.00025 -0.00066
40 4XX -0.15270 -0.26504 0.00022 -0.11315 -0.16789
41 4YY -0.03377 -0.11130 -0.00002 0.00463 -0.14670
42 4ZZ -0.13779 -0.25685 0.00029 0.09344 -0.05785
43 4XY 0.02537 0.04034 -0.00004 -0.05398 -0.03363
44 4XZ 0.00003 -0.00005 -0.05554 -0.00001 0.00006
45 4YZ -0.00001 -0.00008 -0.02912 0.00002 0.00015
46 6 H 1S 0.31391 0.44534 -0.00055 -0.11852 -0.38112
47 2S -0.54622 -0.77798 0.00053 0.43685 -0.99597
48 3PX 0.00452 0.00671 -0.00005 0.09504 -0.04343
49 3PY -0.05127 -0.05236 0.00002 0.04396 -0.13156
50 3PZ -0.00002 0.00002 0.02057 0.00003 -0.00003
26 27 28 29 30
V V V V V
Eigenvalues -- 1.35705 1.39678 1.54381 1.60333 1.80698
1 1 C 1S 0.00002 -0.05921 0.06026 0.00005 -0.00344
2 2S 0.00043 -0.88723 0.66961 0.00042 0.05251
3 2PX 0.00000 -0.03981 0.16042 0.00022 -0.06410
4 2PY 0.00006 -0.03580 -0.00458 0.00018 0.00473
5 2PZ 0.13015 -0.00002 -0.00017 0.03105 0.00004
6 3S -0.00126 3.18984 -3.05485 -0.00257 0.19839
7 3PX 0.00014 -0.66501 0.77967 0.00066 -0.06336
8 3PY -0.00007 0.24874 0.05992 -0.00133 0.59658
9 3PZ -0.33648 0.00042 0.00132 -0.37852 -0.00083
10 4XX 0.00008 0.04648 -0.18060 -0.00027 -0.00431
11 4YY 0.00133 0.15976 0.22927 0.00062 0.25429
12 4ZZ -0.00138 -0.24235 -0.03243 -0.00034 -0.25449
13 4XY 0.00065 -0.35970 -0.27578 -0.00079 -0.18007
14 4XZ 0.55331 0.00042 0.00029 0.03733 -0.00030
15 4YZ 0.26301 -0.00106 -0.00108 0.14617 -0.00103
16 2 H 1S 0.03172 -0.27208 0.25721 -0.09425 0.09969
17 2S -0.19053 -0.35600 0.35130 -0.18035 0.10292
18 3PX -0.19278 0.05389 0.05375 -0.05997 0.20696
19 3PY -0.10187 -0.09044 -0.07625 0.00663 -0.00803
20 3PZ -0.02104 0.08173 0.06880 -0.01008 0.05042
21 3 H 1S -0.03159 -0.27220 0.25661 0.09445 0.09975
22 2S 0.19141 -0.35694 0.34963 0.18026 0.10282
23 3PX 0.19298 0.05369 0.05322 0.06008 0.20596
24 3PY 0.10154 -0.09080 -0.07653 -0.00715 -0.00842
25 3PZ -0.02175 -0.08136 -0.06844 -0.01051 -0.05016
26 4 H 1S 0.00010 -0.24860 0.24169 0.00049 -0.23397
27 2S -0.00022 -0.47419 0.33597 0.00077 -0.19644
28 3PX -0.00019 -0.11817 -0.11659 -0.00017 -0.36113
29 3PY 0.00048 0.06837 0.06779 0.00015 0.13009
30 3PZ 0.18696 -0.00014 -0.00010 -0.01286 -0.00027
31 5 O 1S -0.00001 0.04855 -0.08531 -0.00009 -0.00055
32 2S -0.00034 0.74558 -1.17463 -0.00126 0.05159
33 2PX 0.00001 0.23019 -0.20966 -0.00020 0.04989
34 2PY -0.00002 0.12743 0.12604 0.00001 0.18686
35 2PZ -0.11246 -0.00001 0.00014 -0.05900 -0.00014
36 3S 0.00096 -2.26594 3.46885 0.00361 -0.09701
37 3PX 0.00012 -1.08957 1.03594 0.00109 -0.43960
38 3PY 0.00016 -0.40951 -0.67104 -0.00050 -0.52033
39 3PZ 0.28389 -0.00004 -0.00054 0.32242 0.00045
40 4XX -0.00018 0.39811 -0.45795 -0.00038 -0.02823
41 4YY 0.00037 0.02234 -0.58275 -0.00089 0.30627
42 4ZZ -0.00044 0.23237 0.08789 0.00027 -0.24197
43 4XY -0.00034 0.30901 0.17278 0.00071 -0.46318
44 4XZ -0.39610 -0.00043 -0.00046 0.10855 0.00041
45 4YZ 0.03304 -0.00038 -0.00099 0.73299 0.00018
46 6 H 1S 0.00007 -0.18613 -0.39320 -0.00049 -0.22610
47 2S -0.00018 -0.16737 -0.18108 -0.00026 -0.09781
48 3PX 0.00018 -0.16023 -0.03212 -0.00020 0.34247
49 3PY -0.00009 0.04735 0.01430 0.00012 -0.18432
50 3PZ 0.11330 0.00026 0.00054 -0.42721 -0.00038
31 32 33 34 35
V V V V V
Eigenvalues -- 1.97024 1.98224 2.04134 2.07383 2.08412
1 1 C 1S 0.00005 0.01342 0.00000 0.00773 -0.00003
2 2S -0.00009 -0.16804 0.00001 0.01575 -0.00023
3 2PX 0.00043 0.21039 -0.00001 0.03090 -0.00010
4 2PY -0.00008 -0.02541 0.00001 -0.09421 -0.00008
5 2PZ -0.04402 0.00007 0.00019 0.00009 -0.11334
6 3S -0.00138 -0.12328 -0.00002 -0.30317 0.00130
7 3PX -0.00018 -0.55945 0.00001 0.24091 -0.00052
8 3PY 0.00043 0.08927 -0.00016 0.70207 0.00043
9 3PZ -0.32557 0.00022 -0.00444 -0.00050 0.68534
10 4XX 0.00079 0.73088 -0.00003 -0.01090 0.00036
11 4YY 0.00109 -0.29726 -0.00005 0.47514 -0.00256
12 4ZZ -0.00178 -0.36440 0.00007 -0.45418 0.00220
13 4XY 0.00030 -0.09383 -0.00006 0.39739 0.00005
14 4XZ 0.05287 -0.00017 -0.00004 -0.00053 0.27676
15 4YZ 0.22274 -0.00029 0.00393 -0.00296 -0.57561
16 2 H 1S -0.21094 0.24686 -0.00255 0.30783 0.49449
17 2S -0.00047 0.00306 -0.00012 0.01998 0.00630
18 3PX -0.42022 0.20606 0.04965 -0.10503 -0.14450
19 3PY 0.00177 -0.05523 0.50136 -0.30089 -0.02587
20 3PZ -0.06346 -0.04100 -0.29164 -0.03532 -0.37465
21 3 H 1S 0.21243 0.24653 0.00243 0.30641 -0.49516
22 2S 0.00079 0.00308 0.00011 0.02011 -0.00673
23 3PX 0.42120 0.20502 -0.04974 -0.10540 0.14507
24 3PY -0.00238 -0.05512 -0.50234 -0.30086 0.02514
25 3PZ -0.06353 0.04132 -0.28968 0.03566 -0.37489
26 4 H 1S -0.00046 0.19769 0.00011 -0.59943 0.00058
27 2S 0.00005 -0.00089 0.00001 0.01907 0.00001
28 3PX -0.00038 0.24435 0.00009 0.03435 -0.00046
29 3PY 0.00000 -0.00854 0.00119 -0.37873 -0.00002
30 3PZ -0.00303 0.00000 0.58314 0.00049 -0.29950
31 5 O 1S -0.00001 0.03618 0.00000 -0.01719 0.00007
32 2S -0.00112 -0.35748 0.00004 -0.08822 0.00066
33 2PX -0.00051 -0.39279 -0.00001 -0.00955 -0.00008
34 2PY 0.00011 0.03421 -0.00004 0.05036 -0.00006
35 2PZ -0.14662 0.00017 -0.00106 0.00002 0.05656
36 3S 0.00209 0.05227 -0.00005 0.40070 -0.00216
37 3PX 0.00081 0.23511 0.00003 0.17730 -0.00028
38 3PY -0.00067 -0.07369 0.00008 -0.29148 0.00056
39 3PZ 0.27440 -0.00030 0.00197 0.00024 -0.13854
40 4XX 0.00022 0.30976 -0.00001 -0.07347 0.00017
41 4YY -0.00014 -0.07689 0.00003 0.04873 -0.00016
42 4ZZ -0.00056 -0.24355 -0.00001 -0.07213 0.00053
43 4XY 0.00054 0.00057 0.00002 -0.00456 -0.00069
44 4XZ 0.72909 -0.00090 0.00469 -0.00051 0.02432
45 4YZ -0.22439 0.00027 0.00015 0.00027 -0.00179
46 6 H 1S -0.00066 -0.02770 -0.00002 -0.16893 0.00097
47 2S -0.00020 -0.06594 0.00007 0.08483 0.00001
48 3PX -0.00032 -0.02305 -0.00001 -0.14560 -0.00006
49 3PY 0.00019 -0.01071 -0.00002 0.05584 -0.00017
50 3PZ 0.04900 -0.00006 0.00174 -0.00063 -0.12181
36 37 38 39 40
V V V V V
Eigenvalues -- 2.33795 2.40451 2.53743 2.58752 2.69593
1 1 C 1S 0.02043 0.03020 0.00005 0.04416 0.00007
2 2S 0.16493 -0.08205 -0.00030 -0.15239 -0.00021
3 2PX 0.07420 0.27178 0.00069 0.55622 0.00024
4 2PY 0.00087 -0.00331 -0.00003 0.01529 0.00051
5 2PZ -0.00015 -0.00009 0.12116 -0.00022 -0.15842
6 3S -0.84265 -0.95342 -0.00120 -1.21581 -0.00210
7 3PX 0.35472 0.41660 0.00095 1.04937 -0.00016
8 3PY 0.30768 -0.59157 0.00012 0.14412 0.00210
9 3PZ 0.00139 0.00018 0.80903 -0.00110 -0.24812
10 4XX -0.21312 -0.05028 0.00005 -0.05323 -0.00098
11 4YY 0.18832 0.10765 0.00061 -0.02698 -0.00133
12 4ZZ -0.00359 0.00681 -0.00054 0.10670 0.00260
13 4XY -0.21012 0.40843 0.00028 -0.16386 0.00036
14 4XZ -0.00029 -0.00002 -0.44673 0.00069 0.22979
15 4YZ -0.00042 -0.00052 0.25660 0.00015 -0.09404
16 2 H 1S 0.04150 0.06358 -0.01446 0.03966 0.01501
17 2S 0.19552 -0.13005 0.44641 0.09803 -0.23086
18 3PX -0.15338 0.16519 -0.29859 -0.58296 0.22931
19 3PY 0.04809 0.27026 0.26179 -0.16111 -0.13865
20 3PZ -0.00803 -0.12089 -0.14242 -0.05913 -0.03556
21 3 H 1S 0.04091 0.06344 0.01449 0.03963 -0.01496
22 2S 0.19420 -0.12896 -0.44729 0.09876 0.23344
23 3PX -0.15235 0.16354 0.29866 -0.58345 -0.22773
24 3PY 0.04746 0.27123 -0.26220 -0.16052 0.13460
25 3PZ 0.00654 0.12031 -0.14092 0.06017 -0.03986
26 4 H 1S 0.04380 -0.00670 0.00000 0.06832 0.00004
27 2S -0.14252 0.43561 0.00101 -0.19217 -0.00127
28 3PX 0.28573 -0.54497 -0.00172 -0.32324 0.00093
29 3PY -0.05081 0.22215 -0.00068 0.24217 0.00043
30 3PZ -0.00113 0.00017 -0.54115 -0.00004 0.15495
31 5 O 1S -0.04877 -0.03321 -0.00003 -0.05573 -0.00003
32 2S -0.54157 -0.51549 -0.00053 -0.57269 -0.00075
33 2PX 0.06879 -0.03568 -0.00023 -0.16068 0.00019
34 2PY 0.20159 0.06649 -0.00019 -0.07865 0.00065
35 2PZ 0.00018 0.00003 0.06009 -0.00008 -0.01302
36 3S 1.50969 1.41003 0.00191 2.07355 0.00124
37 3PX 0.06550 0.77331 0.00132 1.04547 0.00133
38 3PY -0.66485 -0.08463 -0.00014 -0.19157 -0.00147
39 3PZ -0.00066 0.00015 -0.48650 0.00048 -0.15596
40 4XX -0.03459 0.01632 0.00042 0.42938 -0.00059
41 4YY 0.36912 0.10433 -0.00056 -0.26895 0.00167
42 4ZZ -0.77548 -0.40610 0.00018 -0.49420 -0.00132
43 4XY 0.44999 0.09984 -0.00078 -0.28359 -0.00139
44 4XZ -0.00069 -0.00026 -0.38213 0.00069 0.51079
45 4YZ 0.00008 -0.00019 0.36210 -0.00025 0.64647
46 6 H 1S -0.87448 -0.24151 0.00044 -0.06418 -0.00075
47 2S 0.08251 0.07762 -0.00036 0.03543 0.00054
48 3PX 0.34356 -0.47543 -0.00092 -0.14590 -0.00177
49 3PY 0.21110 0.39579 -0.00021 -0.12788 0.00183
50 3PZ 0.00017 -0.00072 0.50972 -0.00013 0.90928
41 42 43 44 45
V V V V V
Eigenvalues -- 2.74127 2.80259 2.90207 2.98878 3.24337
1 1 C 1S -0.03193 0.06259 -0.00003 0.00754 -0.10655
2 2S 0.21929 -0.38260 0.00005 0.01464 0.88596
3 2PX -0.18218 0.43125 -0.00024 0.05617 0.54569
4 2PY -0.22400 -0.14489 0.00022 -0.22466 -0.06626
5 2PZ -0.00048 -0.00008 0.03199 0.00030 -0.00011
6 3S 0.64177 -1.21231 0.00085 -0.30211 0.87059
7 3PX -0.02912 0.24271 -0.00036 0.18690 0.29775
8 3PY -0.50255 -0.29476 0.00000 0.02026 -0.01662
9 3PZ -0.00071 -0.00009 0.19929 -0.00008 -0.00014
10 4XX 0.38109 -0.76022 0.00038 -0.05794 -0.73379
11 4YY 0.05324 0.58363 0.00318 -0.57468 0.23215
12 4ZZ -0.54925 0.21881 -0.00344 0.48245 0.30183
13 4XY -0.02779 0.06756 0.00022 0.54295 0.16165
14 4XZ 0.00049 0.00015 0.73202 0.00077 -0.00008
15 4YZ -0.00224 -0.00117 0.56119 0.00331 0.00025
16 2 H 1S -0.01783 0.00692 -0.01536 -0.01244 -0.39877
17 2S -0.31051 0.10342 0.14195 0.02560 -0.35528
18 3PX -0.18724 0.18012 0.39585 0.13187 0.25943
19 3PY 0.30908 0.28409 0.32600 -0.32834 -0.40514
20 3PZ -0.44741 -0.01396 -0.03030 0.20916 -0.60604
21 3 H 1S -0.01789 0.00694 0.01535 -0.01238 -0.39861
22 2S -0.30772 0.10431 -0.14237 0.02561 -0.35508
23 3PX -0.18850 0.17996 -0.39664 0.13012 0.25955
24 3PY 0.31287 0.28486 -0.32416 -0.33021 -0.40267
25 3PZ 0.44592 0.01270 -0.02741 -0.20812 0.60730
26 4 H 1S 0.01438 0.04547 -0.00013 0.13182 -0.39633
27 2S 0.25155 0.36869 0.00019 0.01305 -0.36848
28 3PX -0.27797 0.19640 0.00082 -0.48088 0.33579
29 3PY -0.04064 -0.32458 -0.00153 -0.03879 0.69009
30 3PZ 0.00159 0.00114 -0.65996 -0.00094 -0.00140
31 5 O 1S 0.01425 -0.05493 0.00000 -0.01845 -0.01815
32 2S 0.21301 -0.29692 0.00051 -0.10229 0.03061
33 2PX -0.01018 -0.04940 -0.00016 0.05570 0.08115
34 2PY -0.05597 -0.18510 -0.00040 0.12556 -0.07118
35 2PZ -0.00003 0.00000 0.02278 -0.00001 0.00000
36 3S -0.56426 1.41400 -0.00061 0.25348 0.23453
37 3PX -0.49669 0.83613 -0.00063 0.16304 0.11582
38 3PY 0.34171 0.04319 -0.00020 0.29959 -0.03229
39 3PZ -0.00085 -0.00024 0.06989 0.00029 0.00005
40 4XX -0.07647 0.86605 -0.00056 0.39898 0.20697
41 4YY -0.24020 -0.56614 0.00025 -0.54951 -0.03659
42 4ZZ 0.32310 -0.42386 0.00054 -0.10969 -0.12108
43 4XY 0.43939 0.00303 0.00003 0.40800 0.05310
44 4XZ 0.00189 0.00050 0.25145 -0.00007 -0.00004
45 4YZ 0.00318 0.00090 -0.16758 -0.00080 -0.00008
46 6 H 1S 0.10425 0.02737 -0.00029 0.19296 -0.10589
47 2S -0.02386 -0.10796 -0.00041 0.18963 0.01763
48 3PX 0.48310 0.06250 -0.00067 0.65061 -0.01485
49 3PY -0.25146 -0.38731 -0.00070 0.03725 -0.12713
50 3PZ 0.00396 0.00104 -0.08994 -0.00072 -0.00011
46 47 48 49 50
V V V V V
Eigenvalues -- 3.38616 3.46069 3.54425 3.83667 4.41356
1 1 C 1S -0.00234 0.00004 0.00283 -0.07081 -0.47336
2 2S 0.08794 -0.00045 0.04708 0.55420 2.74169
3 2PX -0.09550 -0.00033 -0.18091 0.09045 -0.07741
4 2PY 0.61864 -0.00424 -0.71486 0.07330 -0.00036
5 2PZ -0.00349 -0.98771 0.00276 -0.00044 0.00003
6 3S -0.23831 0.00052 -0.36216 -1.20662 2.14827
7 3PX 0.03721 -0.00052 -0.01261 0.65494 0.00697
8 3PY 0.38211 -0.00257 -0.42611 -0.17188 0.00086
9 3PZ -0.00186 -0.59013 0.00195 -0.00002 0.00003
10 4XX 0.28579 -0.00058 0.06836 -0.26346 -1.95874
11 4YY 0.21579 0.00218 -0.66994 -0.21758 -2.07548
12 4ZZ -0.52075 -0.00118 0.65854 -0.42882 -2.04930
13 4XY 0.58238 -0.00313 -0.64820 -0.06115 -0.02843
14 4XZ -0.00261 -0.60129 0.00205 -0.00027 0.00002
15 4YZ 0.00046 1.06565 0.00150 0.00005 0.00004
16 2 H 1S 0.30701 -0.74799 -0.29258 0.16958 0.28083
17 2S 0.23323 -0.47025 -0.13685 -0.00011 -0.41707
18 3PX -0.07355 0.27932 0.13632 -0.05431 -0.08362
19 3PY 0.14536 -0.51142 -0.24190 0.07818 0.15866
20 3PZ 0.38813 -0.83435 -0.31552 0.01865 0.26214
21 3 H 1S 0.31030 0.74577 -0.29463 0.17026 0.28081
22 2S 0.23514 0.46885 -0.13857 -0.00005 -0.41710
23 3PX -0.07509 -0.27896 0.13727 -0.05432 -0.08372
24 3PY 0.14632 0.50692 -0.24194 0.07840 0.15765
25 3PZ -0.39245 -0.83377 0.31867 -0.01951 -0.26270
26 4 H 1S -0.45283 0.00201 0.72895 0.11928 0.29839
27 2S -0.28875 0.00131 0.48229 0.04988 -0.40962
28 3PX 0.17112 -0.00102 -0.43456 0.05224 -0.14326
29 3PY 0.64996 -0.00266 -0.83456 0.00140 -0.30823
30 3PZ -0.00123 -0.00653 0.00177 -0.00005 0.00063
31 5 O 1S -0.17090 0.00033 -0.17093 -0.44315 0.07445
32 2S 0.18783 -0.00021 0.05781 -0.12938 -0.04582
33 2PX -0.20798 0.00022 -0.17221 0.01252 0.05253
34 2PY -0.34429 0.00028 -0.24516 0.41093 -0.07531
35 2PZ -0.00001 -0.00468 0.00002 0.00001 0.00000
36 3S 1.74143 -0.00314 1.90528 3.90118 -0.76451
37 3PX -0.39671 0.00032 -0.23229 1.04705 0.01231
38 3PY -0.65677 0.00084 -0.65776 0.21547 -0.08611
39 3PZ 0.00010 0.07494 -0.00018 -0.00002 0.00000
40 4XX -0.63658 0.00083 -0.72700 -1.27414 0.54651
41 4YY 0.09181 0.00058 0.17934 -2.09873 0.31651
42 4ZZ -0.71254 0.00137 -0.79311 -1.40927 0.16106
43 4XY 0.65668 -0.00113 0.35643 -0.50108 0.00005
44 4XZ -0.00028 -0.07965 0.00003 -0.00003 -0.00001
45 4YZ 0.00002 -0.09747 -0.00019 0.00001 -0.00001
46 6 H 1S -0.94092 0.00098 -0.85663 0.92349 -0.14689
47 2S -0.56522 0.00060 -0.56765 -0.11894 -0.02809
48 3PX -0.28837 -0.00021 -0.44874 0.25021 -0.12401
49 3PY -0.92314 0.00107 -0.61970 0.82897 -0.12063
50 3PZ 0.00002 -0.08244 -0.00015 0.00001 -0.00003