Rep:Mod:gxf12345678
NH3
Optimisation Summary
NH3 |
Molecule: NH3
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final Energy E(RB3LYP): -56.55776873 au
RMS Gradient: 0.00000485 au
Point Group: C3V
Optimised N-H Bond Distance: 1.02Å
Optimised H-N-H Bond Angle: 106°
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES
The optimisation file is linked to here.
Vibrations
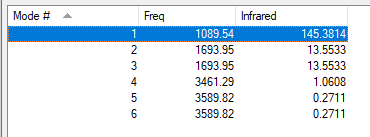
| wavenumber cm-1 |
1090 | 1694 | 1694 | 3461 | 3590 | 3590 |
| symmetry | A1 | E | E | A1 | E | E |
| intensity arbitrary units |
145 | 14 | 14 | 1 | 0 | 0 |
- How many modes do you expect from the 3N-6 rule?
Number of modes = 3(4)-6 = 6
- Which modes are degenerate (ie have the same energy)?
The 2 modes at 1694cm-1 and the 2 modes at 3590cm-1
- Which modes are "bending" vibrations and which are "bond stretch" vibrations?
Bending: 1090cm-1, 1694cm-1
Bond Stretch: 3461cm-1, 3590cm-1
- Which mode is highly symmetric?
3461cm-1
- One mode is known as the "umbrella" mode, which one is this?
1090cm-1
- How many bands would you expect to see in an experimental spectrum of gaseous ammonia?
3 bands
Charge Distribution
Charge on N atom: -1.125
Charge on H atoms: +0.375
It is expected for the N atom to hold a negative charge and the H atoms positive charges, as N is more electronegative and draws the charge density away from the H atoms.
N2
Optimisation Summary
N2 |
Molecule: N2
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final Energy E(RB3LYP): -109.52412868 au
RMS Gradient: 0.00000584 au
Point Group: D*H
Optimised N≡N Bond Distance: 1.105Å
Item Value Threshold Converged? Maximum Force 0.000010 0.000450 YES RMS Force 0.000010 0.000300 YES Maximum Displacement 0.000003 0.001800 YES RMS Displacement 0.000004 0.001200 YES
The optimisation file is linked to here.
Comparison of N≡N Bond Distance in Coordinated Structures
Reported N≡N Bond Distance in Crystal Structure of ICONIA: 1.123Å
Link to Structure: https://www.ccdc.cam.ac.uk/structures/Search?Ccdcid=ICONIA&DatabaseToSearch=Published
The N≡N bond distance in the transition metal complex is longer than the optimised value of 1.105Å. Forming a complex with dinitrogen involves overlap of the non-bonding d-orbitals of the metal with the π* orbital of N≡N. Electrons from the metal flow into the anti-bonding orbitals of the dinitrogen ligand and hence destabilises the N≡N bond[1]. N≡N stretching frequencies in complexes are lower than in the non-complexed dinitrogen molecule and the N≡N is weaker, therefore bond length is larger.
Vibrations
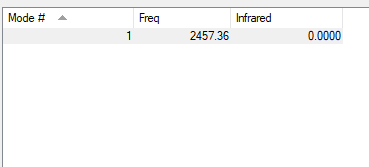
| wavenumber cm-1 |
2457 |
| symmetry | SGG |
| intensity arbitrary units |
0 |
Charge Distribution
Charge on N atoms: 0.000 (linear diatomic)
H2
Optimisation Summary
H2 |
Molecule: H2
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final Energy E(RB3LYP): -1.17853936 au
RMS Gradient: 0.00002276 au
Point Group: D*H
Optimised H-H Bond Distance: 0.743Å
Item Value Threshold Converged? Maximum Force 0.000039 0.000450 YES RMS Force 0.000039 0.000300 YES Maximum Displacement 0.000047 0.001800 YES RMS Displacement 0.000073 0.001200 YES
The optimisation file is linked to here.
Vibrations
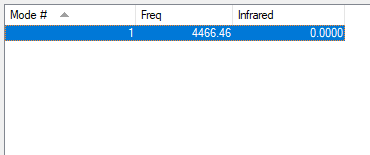
| wavenumber cm-1 |
4466 |
| symmetry | SGG |
| intensity arbitrary units |
0 |
Charge Distribution
Charge on N atoms: 0.000 (linear diatomic)
Haber-Bosch Reaction Energy Calculation for N2 + 3H2 → 2NH3
E(NH3)= -56.55776873 au
2*E(NH3)= -113.11553746 au
E(N2)= -109.52412868 au
E(H2)= -1.17853936 au
3*E(H2)= -3.53561808 au
ΔE = 2*E(NH3) - [E(N2) + 3*E(H2)] = -0.0557907 au = -146.5kJ/mol
The ammonia product is more stable as it is deeper in energy, and it is more favourable to tend towards a lower energy level.
SiH4
Optimisation Summary
SiH4 |
Molecule: SiH4
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final Energy E(RB3LYP): -291.88802760 au
RMS Gradient: 0.00000003 au
Point Group: Td
Optimised Si-H Bond Distance: 1.485Å
Optimised H-Si-H Bond Angle: 109°
Comparison of SiH4 Bond Distance and Angles with Literature
Literature states average bond lengths of Si-H to be 1.48Å[2]. The optimised value of 1.485Å is similar to the literature value. This is expected since SiH4 is a basic molecule and this computational optimisation isolates this molecule from external forces.
The typical H-Si-H bond angle is 109.5°[3]. The optimised value is once again similar to this literature value. The similarity in optimised results is an indicator that the molecule has been optimised properly and correctly.
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES
The optimisation file is linked to here.
Vibrations
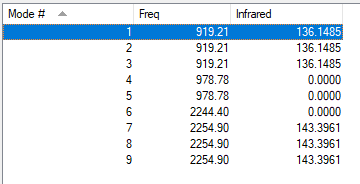
| wavenumber cm-1 |
symmetry | intensity arbitrary units |
diagram | type of vibration[4] |
| 919 | T2 | 136 | 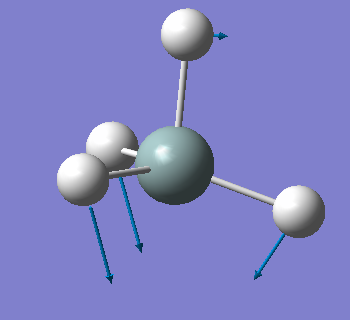 |
bending (asymmetric) |
| 919 | T2 | 136 | 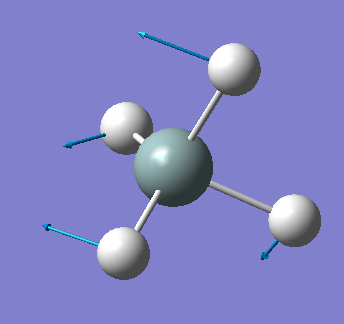 |
bending (asymmetric) |
| 919 | T2 | 136 | 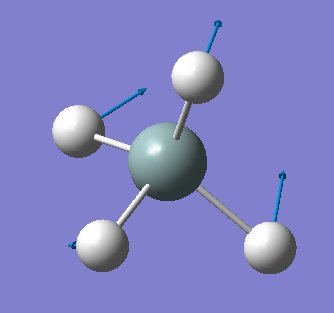 |
bending (asymmetric) |
| 979 | E | 0 | 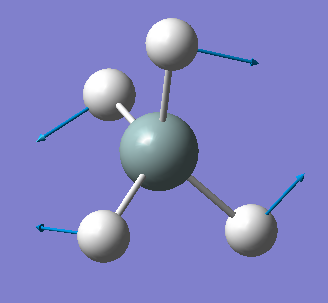 |
bending (symmetric) |
| 979 | E | 0 | 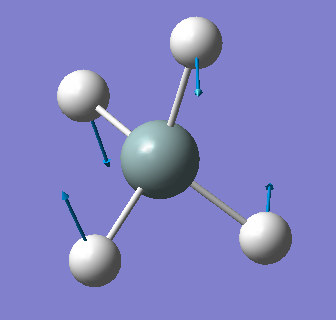 |
bending (symmetric) |
| 2244 | A1 | 0 | 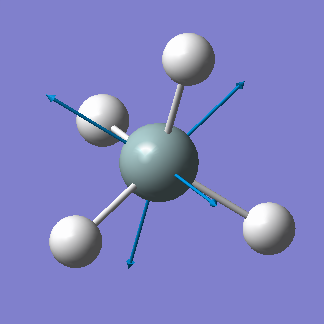 |
stretching (symmetric) |
| 2255 | T2 | 143 | 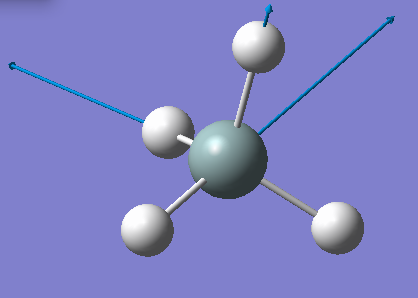 |
stretching (asymmetric) |
| 2255 | T2 | 143 | 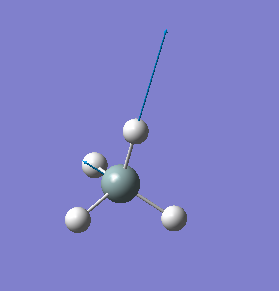 |
stretching (asymmetric) |
| 2255 | T2 | 143 | 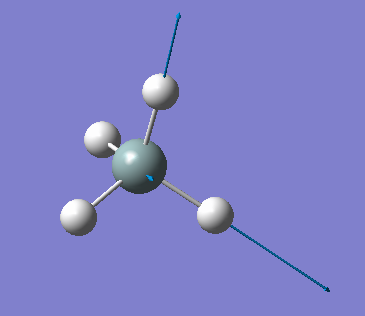 |
stretching (asymmetric) |
Charge Distribution
Charge on Si atom: +0.629
Charge on H atoms: -0.157
Compared to NH3, the central Si atom is now less electronegative than the surrounding H atoms. Electron charge density is now drawn away from Si and hence the H atoms have a negative charge while the charge on Si is positive.
Molecular Orbitals
| Molecular Orbital | Description[5] |
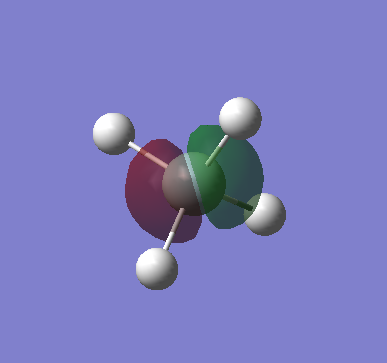 |
This is a fully occupied non-bonding π MO, from an unhybridised 2p AO of Si. There are 3 degenerate MOs of different orientations. As this is in the core shell, it hence has a deeper energy level of -3.64 au. Energy levels are not close enough to that of the H 1s AOs to cause orbital mixing. |
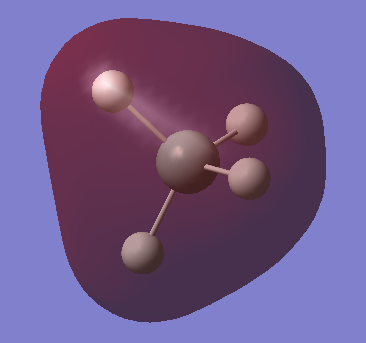 |
This is a fully occupied σ MO, from mixing of Si 3s and H 1s AOs. As this is now in the valence shell, the MO has significantly higher energy (-0.55 au) than the non-bonding MOs from the lower AOs of Si. |
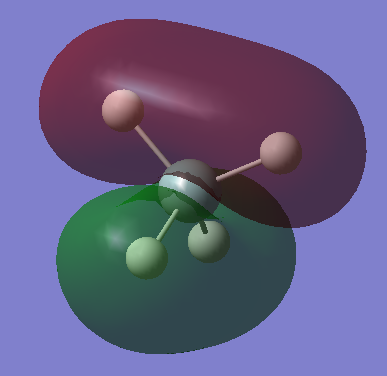 |
This is a fully occupied π MO, from mixing of Si 3p and H 1s AOs. There are 3 degenerate MOs of different orientations. This is in the HOMO region. |
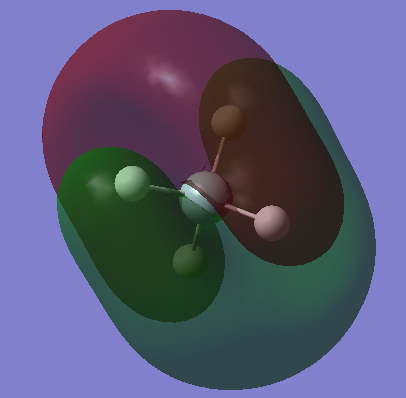 |
This is an unoccupied π* MO, from mixing of Si 3p and H 1s AOs. Similarly to the corresponding bonding π MO, there are 3 degenerate MOs of different orientations. This is in the LUMO region. Filling this MO will destabilise the π bonding MO. |
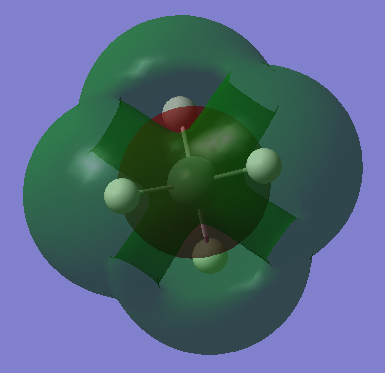 |
This is an unoccupied σ* MO, from mixing of Si 3s and H 1s AOs. This has significantly higher energy than the π* MOs, since the corresponding σ MO is deeper in energy than the π MOs. Filling this MO will destabilise the σ bonding MO. |
References
- ↑ Chatt, J. Molecular nitrogen as a ligand. Pure Appl. Chem. 24, 425–442 (2008).
- ↑ Huheey, pps. A-21 to A-34; T.L. Cottrell, "The Strengths of Chemical Bonds," 2nd ed., Butterworths, London, 1958; B. deB. Darwent, "National Standard Reference Data Series," National Bureau of Standards, No. 31, Washington, DC, 1970; S.W. Benson, J. Chem. Educ., 42, 502 (1965).
- ↑ Liu, K. et al. Quasiclassical trajectory study of H+SiH4 reactions in full-dimensionality reveals atomic-level mechanisms. Proc. Natl. Acad. Sci. 106, 13180–13185 (2009).
- ↑ Gaál-Nagy, K., Canevari, G. & Onida, G. Ab initio calculation of the vibrational modes of SiH 4 , H 2 SiO, Si 10 H 16 , and Si 10 H 14 O . J. Phys. Condens. Matter 20, 224013 (2008).
- ↑ Butler, I. S. A Brief Introduction To Molecular Orbital Theory of Simple Polyatomic Molecules. Quim. Nov. 35, 1474–1476 (2012).
Marking
Note: All grades and comments are provisional and subject to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have received your grade from blackboard.
Wiki structure and presentation 1/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES
NH3 1/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
No
Have you included answers to the questions about vibrations and charges in the lab script?
YES - mostly good answers well done! However there are only 2 visible peaks in the spectra of NH3, due to the low intensity of the other 2 peaks. (See infrared column in vibrations table.)
N2 and H2 0.5/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES
Haber-Bosch reaction energy calculation 1/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 4.5/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
YES - good explanations.
Independence 1/1
If you have finished everything else and have spare time in the lab you could: Check one of your results against the literature, or
YES - well done!
Do an extra calculation on another small molecule, or Do some deeper analysis on your results so far