Rep:Mod:gf609mod1
The Basic Techniques of Molecular Mechanics and Semi-empirical Molecular Orbital Methods for Structural and Spectroscopic Methods By Giovanna Fateh-Iravani
Modelling using Molecular Mechanics
Introduction
Molecular Mechanics is the use of non-quantum mechanical models to express molecular properties. It assumes that the total energy of a molecular system consists of the following:
- Stretch: The sum of all diatomic bond stretches, expressed by a Hookes law potential
- Bend: The sum of all triatomic bond angle deformations, expressed by a Hookes law potential
- Stretch-Bend: A correction that takes account of the coupling between stretching and bending
- Torsion: The sum of all tetra-atomic bond torsions, dependent on the cosine angle
- Van der Waals: The sum of all non-bonded Van der Waals repulsions, using a Lennard Jones potential
- Dipole-dipole: The sum of all electrostatic attractions of individual bond dipoles
The molecular mechanics attempts to minimise the total energy by adjusting bond lengths, bond angles and torsion angles to give the optimised molecular geometry of a conformation. Favourable interactions are shown as negative values. Allinger MM2[1] model implemented in ChemBio3D program will be used.
Diels Alder
Diels alder reactions are an example of a pericyclic reaction between a conjugated diene and a dienophile. The mechanism involves an aromatic transition state with six delocalized π electrons which leads to a stereo specific product.
An example of a Diels Alder 4πs + 2πs cycloaddition reaction is the dimerisation of cylopentadiene forming a norbene system. In this reaction two diastereoisomers are formed under different conditions.[2]
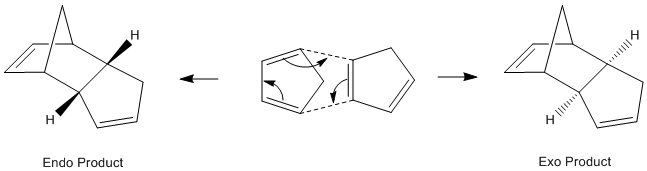
In Literature Baldwin [3] states that the endo isomer is formed preferentially, but at higher temperatures and therefore thermodynamic control, it can be converted to the exo product. The minimum energy conformation was found by molecular modelling and the results were tabulated below:
| Energy in (kcal/mol) | ||
| Exo Product | Endo Product | |
|---|---|---|
| Stretch | 1.2846 | 1.2507 |
| Bend | 20.5799 | 20.8476 |
| Stretch-Bend | -0.8377 | -0.8356 |
| Torsion | 7.6545 | 9.5107 |
| Non-1,4 VDW | -1.4156 | -1.5428 |
| 1,4 VDW | 4.2333 | 4.3193 |
| Dipole/Dipole | 0.3775 | 0.4476 |
| Total Energy | 31.8765 | 33.9975 |
| Total Difference | 2.1210 | |

Under thermodynamic control, although the kinetic product is formed initially, it reverts back to the starting material due to the sufficient energy in the system and so eventually the thermodynamic product dominates. The Exo product is thermodynamically favoured since the 1,4 strain that is present in the endo product is not found in the exo product[4]. The Exo product is therefore less sterically hindered and lower in energy as shown from the results above. The main difference in energy is the Torsion interactions. In the Endo product the Hydrogens are forced closer together and leads to unfavourable interactions. This steric clash means the Endo product has a higher overall energy under these conditions.
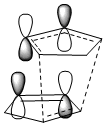
Under Kinetic control there is little thermal energy so the reaction is irreversible[5]. The product with the lower transition state barrier dominates as less energy is required to surmount the barrier and the product with the faster rate of reaction is favoured. In the Diels Alder reaction the Endo product transition state has a through space bonding interaction between the dienophile and the developing π bond at the back of the diene. This stabilising interaction is due to the correct symmetry of HOMO and LUMO frontier orbitals. The Endo transition state is lower in energy than that of the Exo transition state and it is therefore kinetically favoured.
The Hydrogenation of Cyclopentadiene Dimer
From the results shown in the table below it can be seen that Structure 4 has a lower overall energy and is therefore the thermodynamic product. The transition state energy is unknown for both structures so it is unclear which structure is the kinetic product. Structure 4 has lower bending interaction and 1,4 Van der Waals interactions compared to Structure 3 and these more favourable interactions overcome the less favourable torsion interaction in Structure 4.
| Energy in (kcal/mol) | ||
| Structure 3 | Structure 4 | |
|---|---|---|
| Stretch | 1.2780 | 1.0979 |
| Bend | 19.8658 | 14.5234 |
| Stretch-Bend | -0.8349 | -0.5496 |
| Torsion | 10.8068 | 12.4975 |
| Non-1,4 VDW | -1.2257 | -1.0693 |
| 1,4 VDW | 5.6330 | 4.5114 |
| Dipole/Dipole | 0.1621 | 0.1406 |
| Total Energy | 35.6850 | 31.1520 |
| Total Difference | 4.533 | |
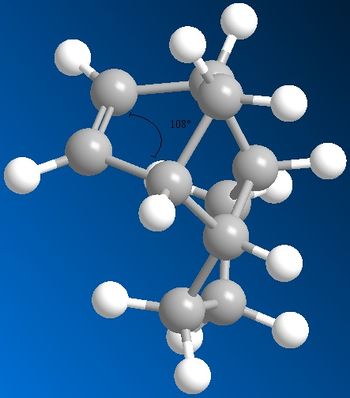
Angular strain is increased as the deviation from the ideal angle increases. Structure 3 has a sp2 bond which ideally would have a geometric angle of 120˚. From modelling it has a angle of 108˚ so a relatively large deviation. Structure 4 has a sp3 bond which modelling shows to be 102˚ this deviation is smaller from the ideal value of 109.5˚ compared to Structure 3. Therefore Structure 4 has less angular strain and so is the most stable product.
 |
 |
Structure 3 also has a double bond which fixes the hydrogen atoms in a geometry that gives unfavourable Van der Waals interactions with the adjacent hydrogen's. In structure 4 this double bond is absent so these hydrogens are able to freely rotate which reduces the interaction. The double bond in structure 4 also fixes the geometry of the hydrogen's, however they are fixed into a favourable site away from adjacent atoms.
Stereochemistry and Reactivity of an Intermediate in the Synthesis of Taxol
Introduction
Taxol is one of the most important organic molecules of today and is an anticancer compound from yew trees.ref clayden . An initial intermediate is used for the total sysnthesis of Taxol and two possible geometries of this intermediate are shown below. In Structure 9 the carbonyl group is pointing upwards in plane with the adjacent hydrogen's of the cyclohexane. In structure 10 the carbonyl group is pointing down. The compound exhibits atropisomerism[6] where on standing it isomerises to the alternative intermediate isomer.
 |
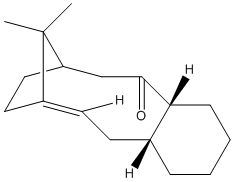 |
MM2 and MMFF94 (Merck Molecular Force Field) energy values were calculated. MMFF94 is a method which uses different parameters [7] to MM2 which have been derived from computational data. The cyclohexyl rings in the Taxol intermediates have two possible conformations, the twist boat and the chair; the chair is more energetically favoured. The table below shows the calculated energies for the two Structure 9 conformations and thwo Structure 10 conformations (one conformations and one twist boat conformation). It can be seen that Structure 10 chair conformation has the lowest energy and is therefore the most stable suggesting that the kinetic and initially formed structure is Structure 9 which on standing converts to the thermodynamic Structure 10.
| Energy in (kcal/mol) | ||||
| Taxol 9 Chair Conformation | Taxol 9 Twist Boat Conformation | Taxol 10 Chair Conformation | Taxol 10 Twist Boat Conformation | |
|---|---|---|---|---|
| Stretch | 2.7853 | 2.8069 | 2.6198 | 2.7074 |
| Bend | 16.5408 | 18.2312 | 11.3364 | 11.8273 |
| Stretch-Bend | 0.4311 | 0.4147 | 0.3432 | 0.3919 |
| Torsion | 18.2412 | 21.1534 | 19.6667 | 22.8862 |
| Non-1,4 VDW | -1.5423 | -1.3036 | -2.1533 | -1.9934 |
| 1,4 VDW | 13.1079 | 14.7646 | 12.8724 | 14.3354 |
| Dipole/Dipole | -1.7244 | -1.6002 | -2.0022 | -1.9969 |
| Total Energy | 47.8397 | 54.4670 | 42.6830 | 48.1580 |
| MMFF94 | 70.5509 | 76.4808 | 60.562 | 66.3285 |
It is stated that the alkene group in the Taxol intermediate reacts slowly and that the hydrogenated intermediates are less stable than Structures 9 and 10. MM2 and MMFF94 energy values were therefore calculated for the hydrogenated Structure 10 for comparison. The total energy values were found to be higher for the hydrogenated molecule than for Structure 10. The bridgehead alkene of Structure 10 is known as a hyperstable alkene[8] which is less strained than the parent hydrocarbon, and therefore has decreased reactivity and increased stability.
| Energy in (kcal/mol) | ||||
| Hydrogenated Structure 10 | ||||
|---|---|---|---|---|
| Stretch | 5.3573 | |||
| Bend | 29.8786 | |||
| Stretch-Bend | 1.2040 | |||
| Torsion | 21.1601 | |||
| Non-1,4 VDW | 1.5635 | |||
| 1,4 VDW | 19.7562 | |||
| Dipole/Dipole | -1.7105 | |||
| Total Energy | 77.2056 | |||
| MMFF94 | 115.812 | |||
The alkene strain energy can be calculated by subtracting the total energy of the alkane from the total energy of the alkene. This gives a negative value of -16.6436 kcalmol-1 for structure 10 which is mainly due to the torsion and Van der Waals components. When comparing the sp2 hybridised alkene angle and the sp3 hybridised alkane angle to ideal bond angles of 120˚ and 109.5˚ respectively, it is clear from the images below that there is a greater difference in the sp3 alkane bond angle suggesting that the alkane is less favourable and therefore the alkene is slow at reacting with electrophilic reagents.
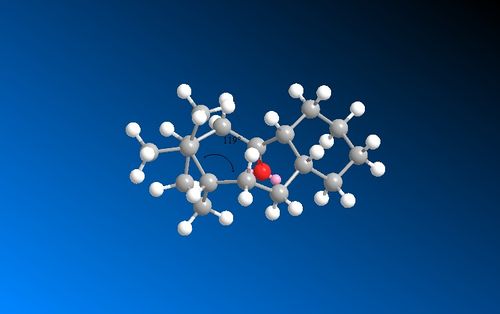 |
 |
As well as torsion strain there is also an added repulsive transannual H-H contact in the alkane which is not present in the alkene, due to a greater number of repulsive H-H Van der Waals interatctions in the alkane. This raises the alkane energy compared to the alkene and is another reason for the hyperstability of the alkene.
Modelling Using Semi-empirical Molecular Orbital Theory
Introduction
MOPAC will be used in this section for calculations as it is designed to implement semi-empirical quantum chemistry algorithms which take into consideration the orbital and electronic contributions of a molecule, which previous calculations above have not.
Regioselective Addition of Dichlorocarbene
Addition of Dichlorocarbene can occur at one of the two alkenes; either the endo alkene (below the chlorine) or the exo alkene (away from the chlorine). The MOPAC/PM6 calculations give a representation of the molecular orbitals of the molecule which will help to predict which alkene addition will occur at. Initially MM2 calculations were carried out to optimise the geometry, results of which are found in the table below:
| Energy in (kcal/mol) | ||
| Molecule 12 | ||
|---|---|---|
| Stretch | 0.6193 | |
| Bend | 4.7362 | |
| Stretch-Bend | 0.0401 | |
| Torsion | 7.6601 | |
| Non-1,4 VDW | 5.7949 | |
| 1,4 VDW | -1.0684 | |
| Dipole/Dipole | 0.1123 | |
| Total Energy | 17.8945 | |
| MOPAC Heat of Formation | 19.7409 Kcal/Mol | |
The molecular orbitals produced from the MOPAC/PM6 calculations are shown below:
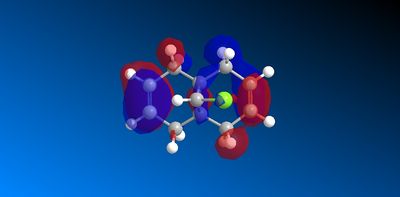 |
 | |
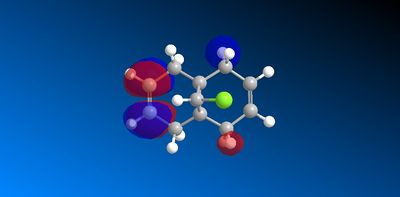 |
 |
|
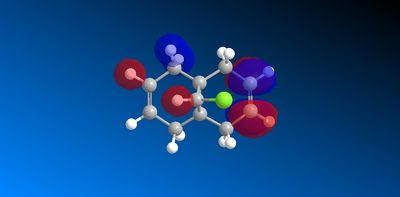 |
The symmetry of the molecule is Cs [9] with a unique σh plane of symmetry shown below, this is consistent with the plane of symmetry found in the molecular orbitals:
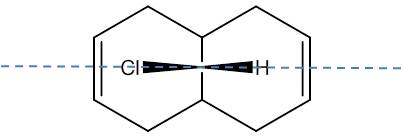
The HOMO has a lower overall orbital energy and greater bonding compared to the LUMO which is evident from the favourable and large interactions. It lies on the endo alkene where the greatest electron density is found. It is therefore the most reactive towards electrophilic attack and therefore addition reactions occur here over the exo alkene. It is also reported [10] that the endo alkene is more nucleophilic than the exo since in the exo there is a stabilizing antiperiplanar orbital overlap between the C-Cl σ* orbital and the occupied exo π orbital. Electron donation from the exo alkene into the antibonding orbital is therefore feasible due to the high electronegativity of chlorine making the C-Clσ* bond low enough in energy for interactions[11]. This results in the exo alkene becoming less electron dense and therefore less nucleophilic.
| Energy in (kcal/mol) | |||
| Mono-ene | With OH Substituent | With CN Sunstituent> | |
|---|---|---|---|
| Stretch | 0.8975 | 0.6066 | 0.6453 |
| Bend | 4.6910 | 5.4372 | 4.5654 |
| Stretch-Bend | 0.0137 | 0.0306 | 0.0312 |
| Torsion | 10.7562 | 7.8008 | 7.4624 |
| Non-1,4 VDW | -1.0632 | -1.4390 | -1.4261 |
| 1,4 VDW | 6.9706 | 4.6918 | 6.7003 |
| Dipole/Dipole | 0.0708 | -0.5589 | -1.2741 |
| Total Energy | 22.3366 | 16.5691 | 16.7045 |
| MOPAC/AM1 | -2.431 | 000000 | 50.289 |
IR Frequency and Vibrational Energies
IR spectrums were predicted for the dialkene, monoalkene, CN substituted dialkene and the OH substituted dialkene:
 |
 |
 |
 |
The IR stretches were analysed in Gauss View, and the vibrations of bonds were looked at. The tabulated results show the stretches that will be focused on in the analysis:
| C=CH Predicted Stretch (cm-1) | C-Cl Predicted Stretch (cm-1) | Exo C=C Predicted Stretch (cm-1) | Endo C=C Predicted Stretch (cm-1) | |
|---|---|---|---|---|
| Dialkene | 689.48 | 770.89 | 1737.26 | 1757.82 |
| Monoalkene | 673.81 | 774.97 | - | 1758.07 |
| OH Substituent | 677.32 | 743.74 | 1753.00 antisymmetric stretch of both alkenes | 1757.76 symmetric stretch of both alkenes |
| CN Substituent | 677.72 | 765.78 | 1706.27 | 1756 |
The results shown that in the monoalkene there is only one C=C stretching frequency corresponding to the endo double bond present which is as expected. The dialkenes exo C=C stretching is slightly lower than that of the endo. This is because electron density on the exo alkene is lost due to the LUMO+2/HOMO-1 interactions which weakens the double bond and therefore lowers yje stretching frequency.
The C-Cl stretching frequency is lower in the dialkene by 4.08cm-1 compared to the monoalkene. This is because the presence of the exo alkene donates electrons into the C-Clσ* orbital which weakens the C-Cl bond decreasing its stretching frequemcy.
When an electron withdrawing group such as CN was substituted onto the exo double bond, the stretching frequency of the C-Cl bond increased. This is because the CN removes electron from the alkene C=C bond reducing the density on the double bond. Therefore there is less donation from the alkene into the C-Clσ* so the C-Cl bond strength increases.
The opposite effect is seen when an electron donating group such as OH is substituted onto the exo alkene. The OH donates electron density into the double bond. Therefore there is increased donation from the double bond to the C-Clσ* orbital which decreases the C-Cl bond strength and the stretching frequency.
Monosaccharide Chemistry: Glycosidation
Introduction
A glycosidation reaction is when a nucleophile replaces a leaving group X on a sugar. The sugars shown below give different anomeric products which are dependent on the orientation of the neighbouring OAc group on the adjacent carbon.
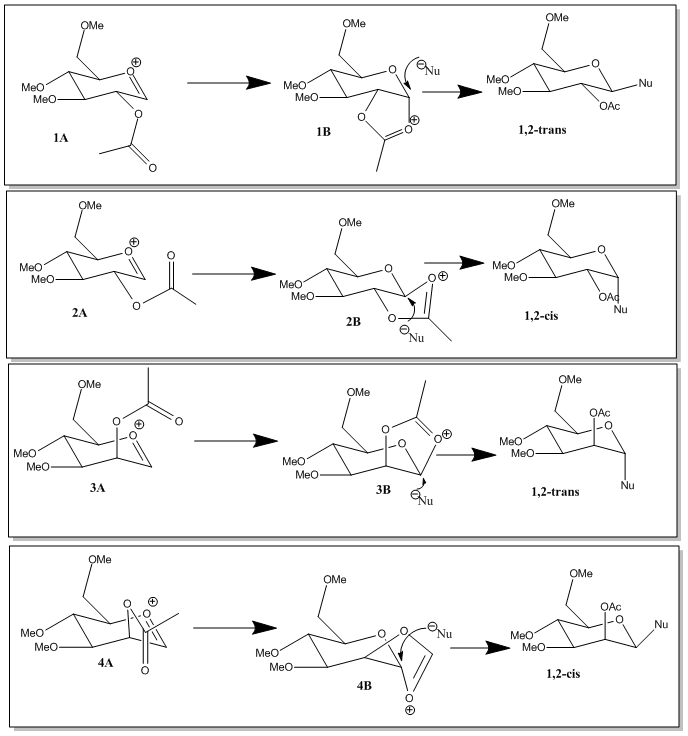
The oxonium cation intermediate is formed when the lone pair on the oxygen is pushed into the ring making X leave. When the intermediate oxoium cation is attacked from the bottom face by the acetyl oxygen, it blocks the bottom face and so the incoming nucleophile replaces it from the top forming the β-anomer. When the acetyl oxygen attacks from the top face, the incoming nucleophile replaces it from the bottom face forming the α-anomer. This makes the mechanism stereospecific.
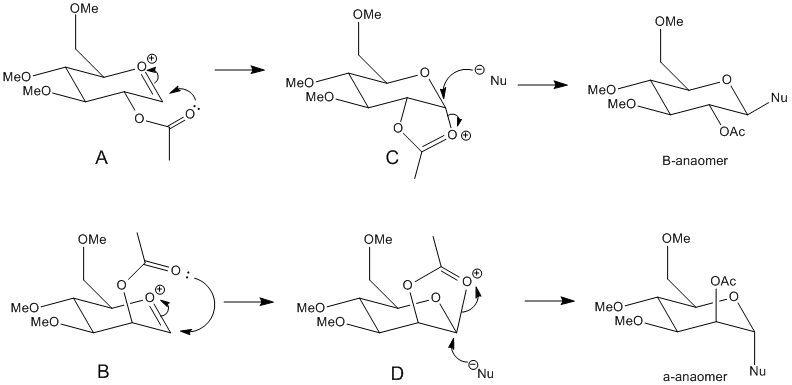
MM2 energies were calculated to optimise the geometries of A, B, C and D shown in the reaction scheme above, and then MOPAC/PM6 energies were calculated. MOPAC/PM6 calculations take into consideration orbital interactions and hydrogen bonding which the MM2 calculations do not. The main difference is that MOPAC/PM6 method thinks of a chemical bond as two electrons in an orbital instead of a rigid bond motif. This must be taken into account since the molecule is complex and contains heteroatoms. The R group that was used in the calculations was Methyl. This was chosen because it is small enough for calculations to be carried out efficiently in a given time. Hydrogen would have also been small enough, however it is not a protecting group and would give rise to added hydrogen bonding interactions.
The MM2 and MOPAC/PM6 energies are shown in the table below. A, B, C and D also have ring flipped isomer versions and so these energies were also calculated and are shown as A’,B’,C’ and D’. The two methods of calculating energies cannot be compared with each other since they take different parameters into consideration, however they are useful for showing a trend.
| Energy in (kcal/mol) | ||||||||
| A MM2 A MOPAC | A' MM2 A' MOPAC | B MM2 B MOPAC | B' MM2 B' MOPAC | C MM2 C MOPAC | C' MM2 C' MOPAC | D MM2 D MOPAC | D' MM2 D' MOPAC | |
|---|---|---|---|---|---|---|---|---|
| Stretch | 2.6013 | 2.3805 | 2.6193 | 2.4769 | 1.9426 | 2.7949 | 1.7953 | 2.9613 |
| Bend | 11.6242 | 12.9009 | 15.1916 | 13.5854 | 12.7858 | 18.4873 | 17.6200 | 18.1804 |
| Stretch-Bend | 0.9771 | 1.0263 | 1.0723 | 0.9643 | 0.6404 | 0.8837 | 0.6943 | 0.8842 |
| Torsion | 3.2245 | 1.5450 | 1.2093 | 0.8173 | 8.0706 | 9.0707 | 8.0611 | 9.2348 |
| Non-1,4 VDW | -0.5847 | -3.1071 | 0.6242 | -1.7798 | -2.7614 | -2.0815 | -3.6667 | -3.0271 |
| 1,4 VDW | 18.8144 | 18.6500 | 17.9583 | 18.3943 | 18.1268 | 19.0520 | 17.9102 | 19.2305 |
| Charge/Dipole | -16.2067 | -0.0061 | -19.7555 | -3.3895 | -8.7590 | -0.3517 | -4.8800 | 0.1245 |
| Dipole/Dipole | 8.3411 | 6.9794 | 8.2973 | 6.2458 | -1.1296 | -1.4455 | -1.0566 | -0.9781 |
| Total Energy | 28.7913 | 40.3689 | 27.2169 | 37.3146 | 28.9161 | 46.4100 | 36.4775 | 46.6106 |
| MOPAC/PM6 | -90.672 | -68.334 | -89.659 | -66.234 | -91.660 | -66.881 | -88.534 | -66.726 |
| Bond interaction distance (angstoms) | 1.6 | 2.9 | 1.6 | 4.5 | 1.6 | 3.8 | 1.6 | 4.2 |
| Angle of Attack (degrees) | 104.8 | 15438 | 105.7 | 149.6 | - | - | - | - |
From the results tabulated it can be seen that the A and B are more stable than their corresponding ring flipped versions - A’ and B’. This is because the in A’ and B’ the carbonyl oxygen of the acetyl group is out of the plane therefore stabilizes the oxonium ion less than in A and B. Therefore the MM2 energies are higher for A’ and B’ and the MOPAC/PM6 energies are less negative ie less stable.
The results also show that A is more stable than B. This is because in A the carbonyl on the acetyl group interacting with the oxonium ion is orientated better to interact therefore the oxonium ion is stabilized better and so the energy is lower compared to B.
From the results C and C’ are lower in energy compared to D and D’. This is explained by the anomeric effect present in C and C’. Here you get stabilisation of the axial substituent since the lone pair from the oxygen in the ring is capable of donationating electrons into the correctly aligned C-Oσ* orbital. Therefore C and C' are more stable and so lower in energy.
Looking at the MOPAC/PM6 energies it can be seen that the energies of A and C are relatively similar and the energies of B and D are also very close in value. This is because MOPAC is capable of determining bonding interactions if the appropriate atoms are close enough in space. The same relationship is less evident in the MM2 energies as MM2 is not capable of this.
The distance between the δ+ carbon on the ring and the carbonyl oxygen were found after MOPAC/PM6 geometry minimisation using ChemBio3D. For A and B the bond distances were less than 2 angstroms which means that both are capable of forming neighbouring group interaction and can therefore stabilise the molecule.
The ring flipped structures A’ and B’ have a distance greater than 2 angstroms between the δ+ carbon on the ring and the carbonyl oxygen therefore there is no neighbouring group interaction to stabilise these structures. The structures are therefore unable to form conformations with the 107o Bürgi-Duntiz[16]. The angles for A’ and B’ have a far greater deviation from this ideal angle than A and B explaining why A and B are more stable.
Mini Project: The Total Synthesis of (-)Cubebol
Introduction
The aim of this project is to show how computational methods of predicting and analysing molecules can be used to distinguish two isomeric products and give the most stable and likely product that will be formed in a synthetic pathway. The reaction pathway below shows the formation of (-)-Cubebol from (-)-Menthone. Many other Cubebol isomers can be formed however the (-)10-epi-cubebol isomer will be the focus of this project.
Cubebol is a natural sesquiterpene alcohol first found in cubeb oil. It has a unique property where it tastes cooling and refreshing.
The energies of all molecules were minimised via MM2 and MOPAC/AM1 methods and the results are tabulated below:
| Energy in (kcal/mol) | |||||
| (-)-menthone (11) | Molecule 17 | Molecule 18 | (-)Cubebol | Isomer | |
|---|---|---|---|---|---|
| Stretch | 0.9088 | 9.9653 | 1.7857 | 1.9917 | 2.0407 |
| Bend | 2.1079 | 74.9425 | 8.7580 | 8.4106 | 8.4897 |
| Stretch-Bend | 0.2182 | -1.3680 | -0.5111 | -0.4770 | -0.4714 |
| Torsion | 7.9488 | 10.4314 | 18.8353 | 18.9947 | 19.4176 |
| Non-1,4 VDW | -2.6108 | -2.4992 | -3.7914 | -4.6315 | -4.2360 |
| 1,4 VDW | 7.4282 | 10.4317 | 8.8816 | 9.5013 | -4.2360 |
| Dipole/Dipole | -1.7110 | 0.1109 | 0.1001 | 0.1421 | 0.1456 |
| Total Energy | 14.2901 | 101.6383 | 34.0691 | 33.9319 | 35.0235 |
| MOPAC/AM1 | -80.299 | -44.960 | -71.43629 | -74.566 | -73.54437 |
From these predicted values Cubebol is the more thermodynamically favoured product compared to the (-)10-epi-cubebol isomer. This is shown by the more negative MOPAC/AM1 value and the lower MM2 energy, implying it is the more stable product.
From the minimised geometries of the two isomers, the NMR spectrum, IR spectrum and Optical rotation were computationally predicted and are shown below. They are compared to literature values[17].
 |
 |
 |
 |
 |
 |
| Carbon Label | Cubebol Predicted Resonance (ppm) | Cubebol Literature Resonance (ppm) | Difference (ppm) | Isomer Predicted Resonance (ppm) | Isomer Literature Resoncance (ppm) | Difference (ppm) |
|---|---|---|---|---|---|---|
| 10 | 78.69 | 80.3 | 1.61 | 78.34 | 80.1 | 1.76 |
| 5 | 45.85 | 44.1 | -1.75 | 44.93 | 42.4 | -2.53 |
| 11 | 41.06 | 39.0 | -2.06 | 43.12 | 41.1 | -2.02 |
| 12 | 37.00 | 36.3 | -0.70 | 36.82 | 36.5 | -0.32 |
| 7 | 35.60 | 33.6 | -2.00 | 35.70 | 33.7 | -2.00 |
| 9 | 35.47 | 33.4 | -2.07 | 35.34 | 32.8 | -2.54 |
| 3 | 32.88 | 31.7 | -1.18 | 33.14 | 30.8 | -2.34 |
| 8 | 32.13 | 30.8 | -1.33 | 31.08 | 29.3 | -1.78 |
| 2 | 31.91 | 29.5 | -2.41 | 30.89 | 29.3 | -1.59 |
| 15 | 29.70 | 27.9 | -1.80 | 29.78 | 28.6 | -1.18 |
| 4 | 28.75 | 26.4 | -2.35 | 26.01 | 20.7 | -5.31 |
| 6 | 27.70 | 22.9 | -4.8 | 23.88 | 19.9 | -3.98 |
| 14 | 22.16 | 20.1 | -2.06 | 22.19 | 19.7 | -2.49 |
| 13 | 21.18 | 19.6 | -1.58 | 21.12 | 19.2 | -1.92 |
| 1 | 19.80 | 18.7 | -1.1 | 19.18 | 17.4 | -1.78 |
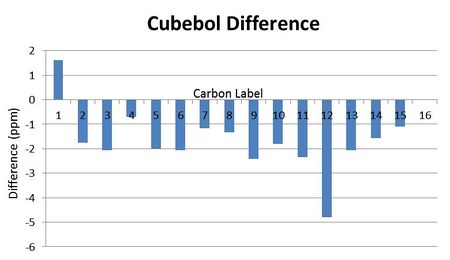
The NMR data is relatively close to that of literature for both isomers, which suggests that the computational predictions are relatively reliable since most fall within the NMR error range of ±5ppm. However there are not any obvious NMR values that can be used to distinguish the two isomers, which was the point of the spectrums. The greatest difference in values is for carbon 6 and carbon 4, however there is no particular reason why these should differ and the error range means these values may overlap regardless. H NMR would be an improved spectrum to predict however these are far more complex to calculate.
Bar charts were made for the difference between the literature values and the computationally predicted values. It is obvious that there is a systemmatic negative error in the results:
 |
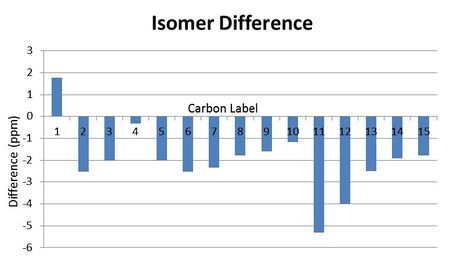 |
IR data for the two isomers is shown below. From looking at the spectrum there is not any peak which is particularly different in the two so again it may be difficult to distinguish the two isomers from this. The main difference in the stretching frequencies is between the CH2 stretches, and this is a possibility for differentiating the two isomers. The CH2 in the molecules are the carbons labelled 3,4,8 and 9 found on both six membered rings. In the isomer both substituents in the first ring are in the same plane which may effect these CH2 groups by decreasing their vibrations since there is more steric bulk. This decrease in vibration would lead to a lower stretching frequencies compared to that of cubebol.
| Cubebol Predicted Stretch (cm-1) | Cubebol Literature Stretch (cm-1) | Difference (cm-1) | |
|---|---|---|---|
| OH Stretch | 3802.2 | 3350 | -452.2 |
| CH Stretch | 2953.5 | 2951 | -2.0 |
| CH Bend | 1501.3 | 1490 | -11.3 |
| CH2 | 1151.7 | 1142 | -9.7 |
| CH | 906.8 | 910 | 3.2 |
| Isomer Predicted Stretch (cm-1) | Isomer Literature Stretch (cm-1) | Difference (cm-1) | |
|---|---|---|---|
| OH Stretch | 3800.6 | 3390 | -410.6 |
| CH Stretch | 3025.8 | 2956 | -69.8 |
| CH Bend | 1498.3 | 1447 | -51.3 |
| CH2 | 949.2 | 946 | -3.2 |
| CH | 934.6 | 931 | -3.6 |
The optical rotation for Cubebol was predicted to be -39.7 deg, this is slightly off that of literature at -48.3 deg, however both values are negative which shows that the direction in which light is polarised has been correctly predicted.
The optical rotation for the isomer was predicted to be -62.75 deg[22], this is slightly off that of literature at -72.3 deg, however they are both negative values which shows that the direction in which light is polarised is correct.
The optical rotation values are useful since they predict the direction in which light will be polarised correctly, however the numerical values are relatively inaccurate so may not be a very useful way to distinguish the two isomers. If the enantiomer of cubebol was investigated instead it would be expected to have a positive optical rotation value, so this technique would be more useful in this instance.
Conclusion
The data collected was reliable since it compared well to literature, however the spectrums would not be a good enough method to distinguish the two isomers.
References
- ↑ Conformational analysis. 130. MM2. A hydrocarbon force field utilizing V1 and V2 torsional terms DOI:10.1021/ja00467a001
- ↑ J. Clayden et al., Organic Chemistry, Oxford University Press, United States, 2001, pp 484
- ↑ Baldwin, J. E., Rules for Ring Closure, J. Chem. Soc., Chem. Commun. 1976, 734. doi:10.1039/C39760000734
- ↑ J. Clayden et al., Organic Chemistry, Oxford University Press, United States, 2001, pp 484
- ↑ J. Clayden et al., Organic Chemistry, Oxford University Press, United States, 2001, pp 484
- ↑ 3rd Year Computational Chemistry Online Lab Manual, Module 1, 2011
- ↑ Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94, Thomas A. Halgren, J. Comp. Chem.; 1996; 490-519, doi:10.1002/(SICI)1096-987X(199604)17:5/6<490::AID-JCC1>3.0.CO;2-P
- ↑ W. F. Maier et al., J. Am. Chem. Soc., 1981, 103, pp 1891
- ↑ T. Hunt, 2nd Year Inorganic Chemistry Lecture Course - Molecular Orbitals, Character Table, 2010
- ↑ 3rd Year Computational Chemistry Online Lab Manual, Module 1, 2011
- ↑ B. Halton et al., J. Chem. Soc., Perkin Trans 2, 1992, pp 447
- ↑ http://hdl.handle.net/10042/to-11813
- ↑ http://hdl.handle.net/10042/to-11818
- ↑ http://hdl.handle.net/10042/to-11822
- ↑ http://hdl.handle.net/10042/to-11821
- ↑ A. Spivey, 3rd Year Organic Lecture Course - An Introduction To Stereoelectronics, Lecture 3, 2011
- ↑ DOI:10.1021/jo9022974
- ↑ http://hdl.handle.net/10042/to-11837
- ↑ http://hdl.handle.net/10042/to-11832
- ↑ http://hdl.handle.net/10042/to-11829
- ↑ http://hdl.handle.net/10042/to-11835
- ↑ http://hdl.handle.net/10042/to-11759
