Rep:Mod:feihe
Module 1: Organic
Introduction
This module contains two parts. One part containse 5 exercise which is solved by using different programmes. The other part is to do a mini-research project which is often seen in chemistry synthetic lab.
Experimental
Exercise 1: The hydrogenation of Cyclopentadiene Dimer
Cyclopentadiene will dimerise to form 2 dimers, one is endodimer and the other is exodimer. And the hydrogenation of these dimers will give dihydro derivatives.
Now we need to get the final energies of the dimers and compare the energy difference between the 2 dimers. And we can obtain the energies of the dimers by MM2 force field option of Chem 3D programme. By applying the programme, we get the following datas.
 |
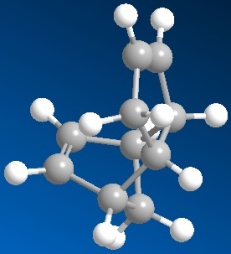 |
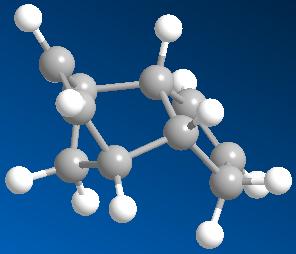
| 3D structure of exodimer (dimer 1) | structure of endodimer (dimer 2) |
| Energy: 31.8834 kcal/mol | Energy: 34.0153 kcal/mol |
From the data obtained above, we can see that the final energy of the dimer 1 is about 3 kcal/mol smaller than that of dimer 2. So dimer 1 is less strained than dimer 2 and this means that exodimer is more hindered than endodimer. So endodimer 2 should be produced in the cyclodimerisation reaction in a thermodynamic sense.
From the dimerisation of the cyclopentadiene, we know that endodimer 2 is experimentally more favourable product than exodimer1. This means that the cyclodimerisation of cyclopentadiene is under kinetic control since the transition state of endodimer 2 is more stable that of exodimer 1.
The hydrogenation of the dimer 2 will give dihydro derivatives 3 or 4.
By applying the MM2 of the Chem 3D programme, we can get the stretching, bending, torsion, van der waals and hydrogen bonding energy terms of the derivative 3 and 4. And we can get the total energies of the 2 derivatives as well.
 |
 |
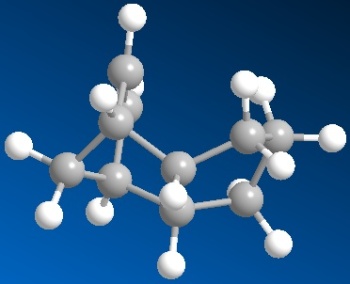
| 3D structure of dihydro derivative 3 | 3D structure of dihydro derivative 4 |
| Total Energy: 35.9337 kcal/mol | Total Energy: 31.1540 kcal/mol |
| All terms are in kcal/mol unit. | Derivative 3 | Derivative 4 |
|---|---|---|
Stretching |
1.2324 | 1.0963 |
Bending |
18.8641 | 12.4972 |
Torsion |
12.2469 | 12.4972 |
Van der waals |
5.7523 | 4.5124 |
Dipole-dipole |
0.1631 | 0.1407 |
By comparing the total energies of the 2 derivatives, we can see that derivative 3 is in higher total energy than derivative 4. This means that derivative 3 is more strain than derivative 4, so it is less favoured in the hydrogenation of the dimer in thermodynamic sense. So derivative 4 is less hindered and therefore the double bond in the cyclohexene ring in endodimer 2 eases the hydrogenation.
From the above table, we can see that bending and torsion terms contribute mostly in the total energy. All the energy terms are higher in energy in derivative 3 and those in derivative 4 apart from the torsion term. But the difference in torsion energies in 2 derivatives is quite small and derivative 3 is still less favoured. As sum up, the hydrogenation of the dimmers is under thermodynamic control.
Exercise 2: Stereochemistry of Nucleophilic additions to a pyridinium ring (NAD+ analogue)
There are 2 reactions we need to explore in this exercise.
1st reaction is following:
Mechanism of 1st reaction:
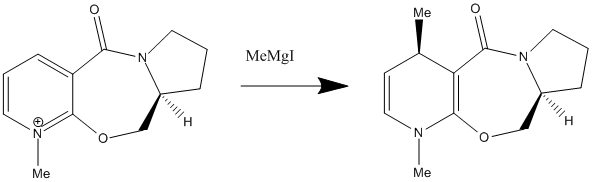
In the 1st reaction, MeMgI is a Grignard reagent which is nucleophilic carbon species. . Since carbon 2,4,6 positions are electrophilic, Grignard reagent will attack these carbon position in pyridine ring. But carbon 4 position is more electrophilic than the other positions. So the Methyl group with negative charge would mostly like to attack the carbon 4 position in the pyridine ring in order to stabilise the N atom with positive charge. This is known as 1,4-addition (major reaction) and it is a quite stereoselective, regioselective and major addition. In this case, 1,6-addition is a minor addition. The major distereoisomer formed in the 1,4-addition reaction has got the methyl group orientated anti to the H atom bonded to the chiral centre of the L-prolinol auxiliary.
2nd reaction is following:
Mechanism of reaction 2:
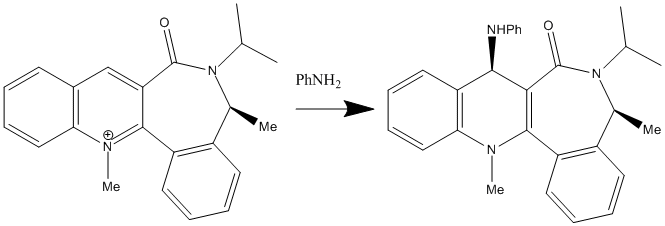
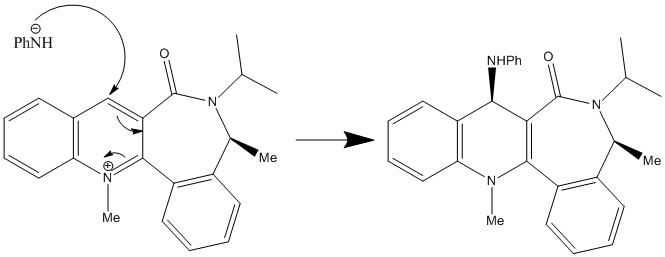
In the 2nd reaction, the PhNH2 is acted as a nucleophile, so it would like to attack the electrophilic carbon position 4 only in this case due to hindered reason. The orientation of the NHPH of the product is due to the steric control of the C=O lactam. And the C3-C=O bond is the most configurationally element controls the stereo selectivity of the addition.
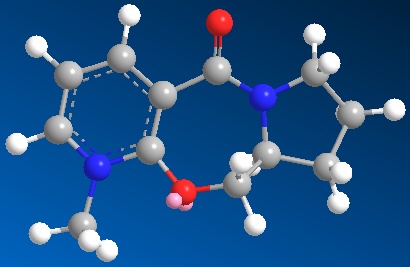
First of all, we need to draw out the structures of the reactant 5 and 7 by using Chem Draw programme. Copy the structures and paste into the Chem 3D programme. By applying the MM2 of the Chem 3D programme, we minimise the models produced in the 3D programme and can get the total energies of the pyridinium reactant 5 and pyridinium reactant 7.
 |
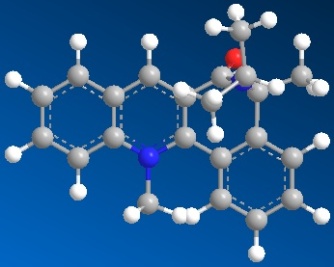 |
| 3D structure of active prolinol derivative 5 | 3D structure of active prolinol derivative 7 |
| Total energy: 26.3148 kcal/mol | Total Energy: 32.0338 kcal/mol |
Since the derivative 7 has got more double bonds and this feature will give the molecule quite a lot of strain, hence the total energy of derivative 7 is a little bit higher than that of derivative 5. I have tried to do several starting points for my optimization and the above total energies are the minimums of those values. For the derivative 5, if the starting point is C6 in the pyridine ring (try to pull the C6 atom a bit away from the ring), then the total energy is 26.3747 kcal/mol and if the starting point is C5 in the pyridine ring (try to pull the C5 atom inside the ring), then the total energy is still 26.3747 kcal/mol. All the other atoms acted as starting point will give higher values than 26.3148 value. For derivative 7, if the starting point is the carbon in the carbonyl group (away from the original position), this will give the value of 33.5565 which is higher than the above 32.0338 value. So the minimum energy values obtained are 26.3148 for derivative 5 and 32.0338 for derivative 7.
From the two 3D models, we can see that the carbonyl group is in the same phase with respect to the ring in derivative 5 whereas the carbonyl group is not in the ring phase. Now from the 3D structures, we can get the angles of the carbonyl group with respect to pyridine aromatic ring.
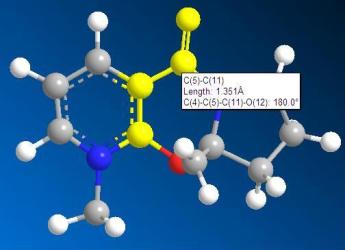 |
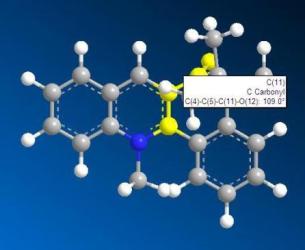 |
| angle of derivative 5: 180.0 degree | angle of derivative 7: 109.0 degree |
Now apply the same method above for molecule 6 and molecule 8 and give the following 3D structures.
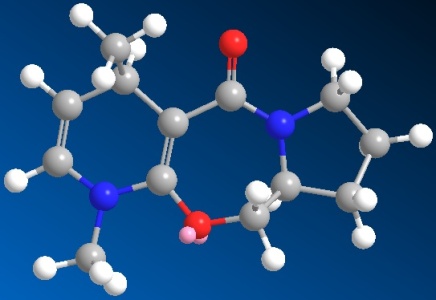 |
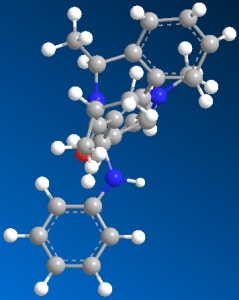 |
| 3D structure of molecule 6 | 3D structure of molecule 8 |
| Total energy: 28.8693 kcal/mol | Total Energy: -7.9114 kcal/mol |
Get the dihedral angle of the 2 models of the molecule 6 and molecule 8.
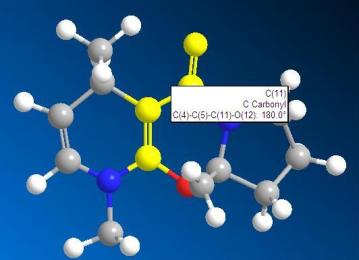 |
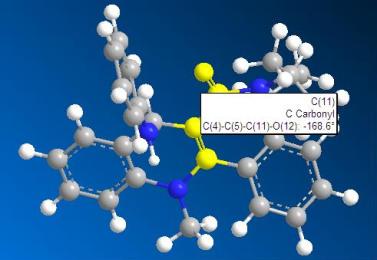 |
| angle of molecule 6: 180.0 degree | angle of molecule 8: 168.8 degree |
From the above 2 angle graphs, we can see that the carbonyl group in molecule 6 is in the same phase with respect to the pyridine ring whereas the carbonyl group in the molecule 8 is not in the same phase with the pyridine ring.
Table of dihedral angle of the 4 molecules:
| Molecule 5 | Molecule 6 | Molecule 7 | Molecule 8 | |
|---|---|---|---|---|
| angle/degree | 180.0 | 180.0 | 109.0 | 168.8 |
From the above table and the 3D models of the 4 molecules, we can see that molecule 5 and 6 are in the same phase with the pyridine ring whereas molecule 7 and 8 are point into the paper. So the NHPh group will point out of paper due to steric hinder reason and it doesn’t like to get close to the C=O bond since both NHPh and C=O are electron rich and they will repel each other. For molecule 5 and 6, the methyl group would like to point out of the paper because of the H atom bonded to the chiral centre of the L-prolinol auxiliary. The methyl group and H atom would like to be anti with each other due to repulsion reason.
Exercise 3: Stereochemistry and Reactivity of an Intermediate in the Synthesis of Taxol
Draw the 2 models of the isomer 10 and isomer 11 and run the MM2 of the Chem 3D programme to get the total energies of the 2 models. Then compare the energies of the 2 isomers and hence determine which is more stable. Therefore, we can know that the stereochemistry of carbonyl addition depends on which isomer.
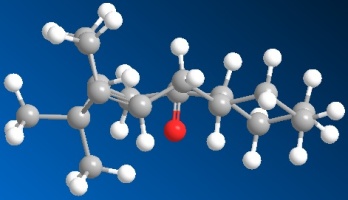 |
| 3D Structure of resulting optimised isomer 10 |
| Total energy: 176.0327 kcal/mol |
For isomer 10, just paste the Chem draw molecule into the Chem 3D programme firstly, this will give a total energy of 176.0327 kcal/mol. Then I tried several times to minimise the energy and I found out the energy is a much smaller if I tried to let the structure of the molecule more likely to be in chair-chair conformation. If I try to pull the oxygen atom away from the original position, the total energy will be larger. And the minimum energy of the isomer 10 among all the attempts found is 44.8312 kcal/mol. And after minimise the structure of the isomer 10, the resulting optimised structure has got the C=O group pointing down. And the whole structure is similar to the chair-chair form.
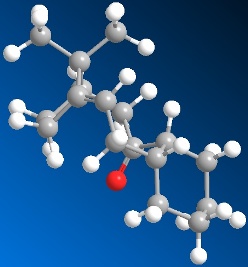 |
| 3D Structure of resulting optimised isomer 11 |
| Total energy: 98.3674 kcal/mol |
For isomer 11, firstly paste the Chem draw molecule to the 3D programme and run the MM2 force field option and get the total energy of isomer 11. And the first attempt total energy is 98.3674 kcal/mol which is much lower than the first attempt energy of isomer 10. Change the structure similar to the chair-chair form and get the smaller value of energy. After several attempts, I got the minimum energy of those values is 44.2970 kcal/mol. After stabilise the structure of the isomer 11, the resulting optimised structure has got the C=O group pointing down. And the whole structure is similar to the chair-chair form.
By comparing the 2 energies of the isomers, the energy of isomer 11 is smaller than that of isomer 10. So the isomer 11 is more stable. Therefore, the stereochemistry of carbonyl addition depends on isomer 11. This is due to the bridged E-cyclononenone. In isomer 10, the bridged E-cyclononenone in the central position is efficient rigidified to lock the fused cyclohexane which is adjoined with the E-cyclononenone into a very energetic twist-boat conformation. And this problem is improved in the chair structure of isomer 11, the non-bonded transannular interactions within the ring core of structure 11 are heightened. And the lone pairs of the oxygen atom will have dipole-dipole repulsion with the methyl group in the bridge of the boat structure. So chair form is preferred. Therefore, the isomer 11 is more stable than isomer 10.
The bridging will give a lot of strain to the both conformations. So both conformations have a lot of strain and it is hard to relieve the steric strain due to steric reason. And the H atom in the double bond will like to H-bond with the O atom in the C=O bond. So it is hard to take off the H atom and it is hard to do addition to the double bond due to steric reason. So the alkene reacts slowly.
Exercise 4: How one might induce room temperature hydrolysis of a peptide
There are 2 reactions we concerned in this exercise.
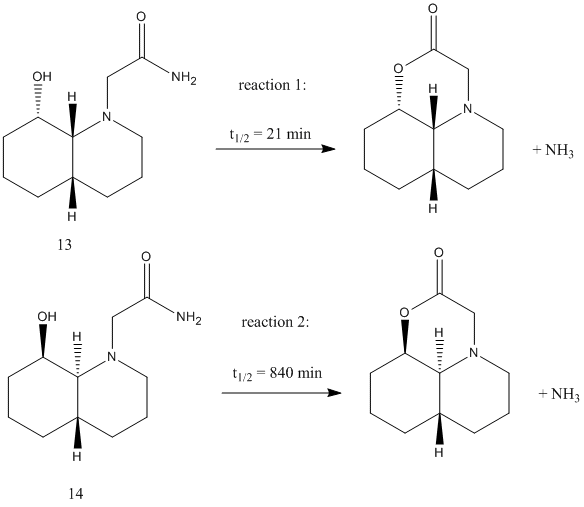
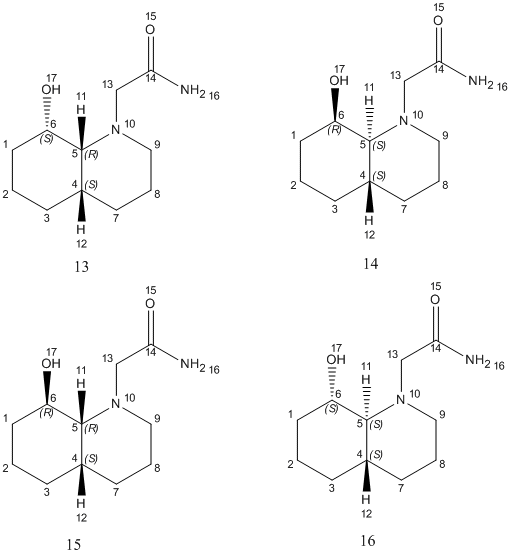
The name of the isomer 13 is (4aα,8β,8αα)-1-ethylamido-8-hydroxydecahydroquinoline and the name of the isomer 14 is (4aα,8α,8αβ)-1-ethylamido-8-hydroxydecahydroquinoline. The isomer 13 is a cis-isomer whereas isomer 14 is a trans-isomer. Both isomers has got a lot of constrain but there is no steric compression and strain between the –OH group and the amide group. So the hydrolysis of the 2 groups can occur in both reactions and it will take much less than 500 years.
Draw out the structures of the diastereomers of the isomer 13 and isomer 14.
The cis-isomer 13 has got 2 chair-chair conformations, one has the hydroxyl group equatorial and one has the hydroxyl group axial. The equatorial one is more stable than the axial one since the –OH group in the axial one is involved in synaxial interations. The trans-isomer 14 has only 1 chair-chair conformation. So for the isomer 13, we are going to use the equatorial hydroxyl group and for isomer 14, we are going to use the axial hydroxyl group.
Build the models of the above 4 conformations (2 conformations for each isomer): Here the one isomer has got 2 conformations and the conformations depend on the position of the ethylamido group. The major conformation of each isomer has the ethylamido group in equatorial position and the minor conformation of each isomer has the group in axial position.
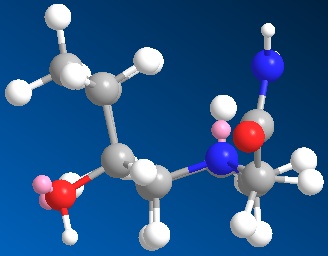 |
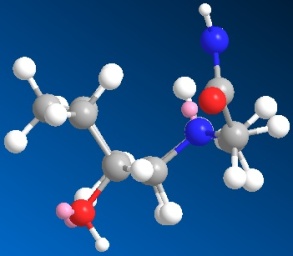 |
| 3D optimised structure of isomer 13 | 3D optimised structure of isomer 13 |
| (major conformation) | (minor conformation) |
| Total Energy: 16.9656 kcal/mol | Total Energy: 16.9835 kcal/mol |
 |
 |
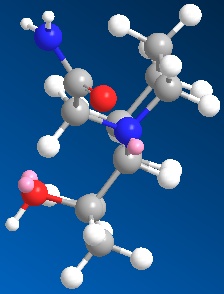
| 3D optimised structure of isomer 14 | 3D optimised structure of isomer 14 |
| (major conformation) | (minor conformation) |
| Total Energy: 20.0563 kcal/mol | Total Energy: 9.6723 kcal/mol |
From the optimised structure of isomer 13, we can see that the ring N-substituent is oriented axial with respect to the decalin ring. And from the optimised structure of isomer 14, the ring N-substituent is oriented equatorial with respect to the decalin ring. For major conformation of isomer 14, the OH group cannot approach the carbonyl of the peptide bond, however, the OH group can approach in the minor conformation of isomer 14.
| Conformation | Major of isomer 13 | Major of isomer 13 | Major of isomer 14 | Major of isomer 14 |
|---|---|---|---|---|
| Total energy/ kcalmol^-1 | 16.9656 kcal/mol | 16.9835 kcal/mol | 20.0563 kcal/mol | 9.6723 kcal/mol |
From the above table, we can see that for the isomer 13, both conformations are quite similar in energy whereas the 2 conformations of isomer 14 are very different in energy. Both minor conformations of both isomers have lower energies than those of major conformations so they are more stable. So they are hardly likely to react. So the major conformations would like to be hydrolysed. Kinetic analysis depends on the initial rates of the reactions, i.e whether hydroxyl group can approach the carbonyl of the peptide or not. For the major conformation of isomer 13, the hydroxyl group can approach. For the minor conformation of isomer 14, the hydroxyl group approach. So for isomer 14, the conformation firstly will change to the minor conformation with lower energy and this is not a rapid inversion. So the initial rate of the hydrolysis for isomer 14 will be slow.
See the following amide cleavage in isomer 13 and isomer 14.
For isomer 13:
For isomer 14:
The major conformations of both isomers have got the ethylamido group in equatorial position. In the amide cleavage in isomer 13, the equatorial ethylamido group could attack the hydroxyl group by intramolecular nucleophilic attack. However, in the amide cleavage in isomer 14, the equatorial ethylamido group could not attack the hydroxyl group due to its position. So the ethylamido group would like to change from equatorial position to axial position to attack the hydroxyl group. And this will cost energy, so the hydrolysis reaction will be slow for isomer 14.
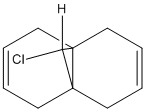
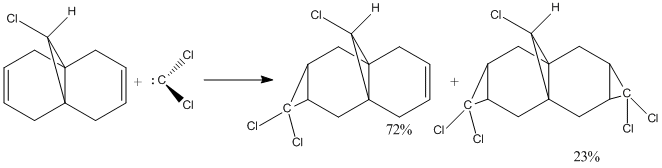
Exercise 5: Regioselective Addition of Dichlorocarbene
The aim of this exercise is to use different methods to calculate the energy and geometry of the follow molecule 12.
Procedure:
First method: use the MM2 option of the Chem 3D programme to predict the structure of molecule 12.
And I got the following information about this molecule:
MM2 Minimization------------
Warning: Some parameters are guessed (Quality = 1).
Iteration 114: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 0.6244 Bend: 4.6971 Stretch-Bend: 0.0398 Torsion: 7.6930 Non-1,4 VDW: -1.0711 1,4 VDW: 5.8011 Dipole/Dipole: 0.1126
Total Energy: 17.8970 kcal/mol Calculation completed
Second method: use the HF/STO-3G method to predict the structure.
However, when I tried to run this method, it produced an error. Firstly, I thought there is something wrong with the molecule I drew by using the Chem Draw programme. And I redrew the whole molecule. And I try to run this method again. But it had the same problem as before. And I double checked the procedure and it is followed by the lab script.
See the below error details:
See the end of this message for details on invoking just-in-time (JIT) debugging instead of this dialog box.
- Exception Text **************
System.UnauthorizedAccessException: Access to the path 'C:\gchk6335.chk' is denied.
at System.IO.__Error.WinIOError(Int32 errorCode, String maybeFullPath) at System.IO.File.InternalCopy(String sourceFileName, String destFileName, Boolean overwrite) at CambridgeSoft.ChemOffice.GaussianDriver.Driver.RegenerateFChk() at CambridgeSoft.ChemOffice.GaussianDriver.Driver.ReadResults(CEXML results, Int64 outputStartPos) at CambridgeSoft.ChemOffice.GaussianDriver.Driver.FinalResults(CEXML results) at CambridgeSoft.ChemOffice.DriverShared.TDriver.OnComputeResults(CEXML& results) at CambridgeSoft.ChemOffice.GaussianDriver.Driver.CambridgeSoft.ChemOffice.COEA.IDriver.OnComputeResults(CEXML& ) at CambridgeSoft.ChemOffice.COEA.WorkThread.GetResults() at CambridgeSoft.ChemOffice.COEA.WorkThread.End(Boolean getResult) at CambridgeSoft.ChemOffice.COEA.WorkThread.TimerProc(Object source, EventArgs e) at System.Windows.Forms.Timer.OnTick(EventArgs e) at System.Windows.Forms.Timer.TimerNativeWindow.WndProc(Message& m) at System.Windows.Forms.NativeWindow.Callback(IntPtr hWnd, Int32 msg, IntPtr wparam, IntPtr lparam)
- Loaded Assemblies **************
mscorlib
Assembly Version: 2.0.0.0 Win32 Version: 2.0.50727.1433 (REDBITS.050727-1400) CodeBase: file:///C:/WINNT/Microsoft.NET/Framework/v2.0.50727/mscorlib.dll
CambridgeSoft.ChemOffice.COEA
Assembly Version: 11.0.0.1 Win32 Version: 11.0.0.574 CodeBase: file:///C:/WINNT/assembly/GAC_MSIL/CambridgeSoft.ChemOffice.COEA/11.0.0.1__b1703fdac613d3b0/CambridgeSoft.ChemOffice.COEA.dll
System
Assembly Version: 2.0.0.0 Win32 Version: 2.0.50727.1433 (REDBITS.050727-1400) CodeBase: file:///C:/WINNT/assembly/GAC_MSIL/System/2.0.0.0__b77a5c561934e089/System.dll
System.Windows.Forms
Assembly Version: 2.0.0.0 Win32 Version: 2.0.50727.1433 (REDBITS.050727-1400) CodeBase: file:///C:/WINNT/assembly/GAC_MSIL/System.Windows.Forms/2.0.0.0__b77a5c561934e089/System.Windows.Forms.dll
System.Drawing
Assembly Version: 2.0.0.0 Win32 Version: 2.0.50727.1433 (REDBITS.050727-1400) CodeBase: file:///C:/WINNT/assembly/GAC_MSIL/System.Drawing/2.0.0.0__b03f5f7f11d50a3a/System.Drawing.dll
CambridgeSoft.ChemOffice.ChemPropPro
Assembly Version: 11.0.0.1 Win32 Version: 11.0.0.574 CodeBase: file:///C:/Program%20Files/CambridgeSoft/ChemOffice2008/Common/COEA/CambridgeSoft.ChemOffice.ChemPropPro.dll
CambridgeSoft.ChemOffice.DriverShared
Assembly Version: 11.0.0.1 Win32 Version: 11.0.0.574 CodeBase: file:///C:/Program%20Files/CambridgeSoft/ChemOffice2008/Common/COEA/CambridgeSoft.ChemOffice.DriverShared.DLL
CambridgeSoft.ChemOffice.CoreChemistry
Assembly Version: 11.0.0.574 Win32 Version: 11.0.0.574 CodeBase: file:///C:/Program%20Files/CambridgeSoft/ChemOffice2008/Common/COEA/CambridgeSoft.ChemOffice.CoreChemistry.DLL
CambridgeSoft.ChemOffice.ChemPropStd
Assembly Version: 11.0.0.1 Win32 Version: 11.0.0.574 CodeBase: file:///C:/Program%20Files/CambridgeSoft/ChemOffice2008/Common/COEA/CambridgeSoft.ChemOffice.ChemPropStd.dll
CambridgeSoft.ChemOffice.CLogPDriver
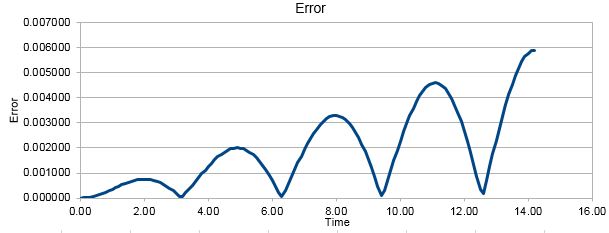
The following the error graph:
Mini Project
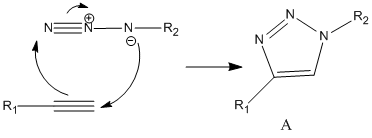
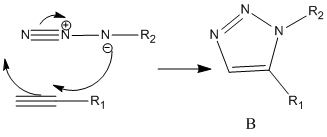
Aim: This mini project is focusing on the click reaction. And we want to use some spectroscopical methods to differentiate the isomeric products.
Click Reaction:
 Where A and B are regioisomeric products.
Where A and B are regioisomeric products.
Mechanism of the reaction:
Procedure:
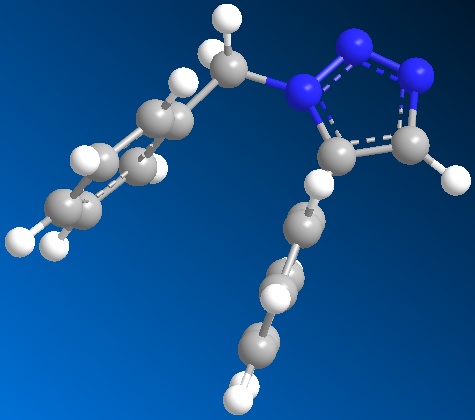
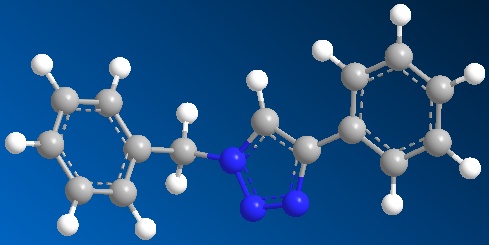
1. Copy down the example in Table 1, entry 1 of the reference paper. And draw out its structure by using Chem Draw programme and draw out its isomer as well. And paste the structures into Chem 3D programme and hence obtain the models of isomer A and isomer B. Then run the MM2 option to refine the geometry of the models.
2. The Gaussian input files of both models are created and edited followed by the lab script.
3. 2 files are then submitted to the SCAN for determine the final optimised geometry for about 3 hour.
4. Download the finished files and edit the file. Then resubmit the files to the SCAN for NMR chemical shift calculation. Then can get the files we needed.
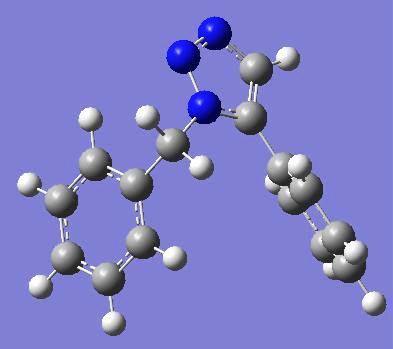
Optimised model for isomer A by using SCAN:
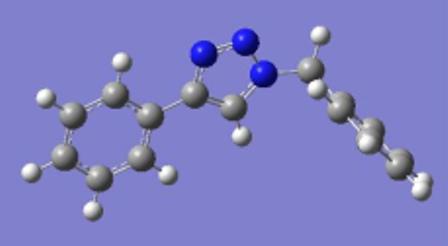
Optimised model for isomer B by using SCAN:
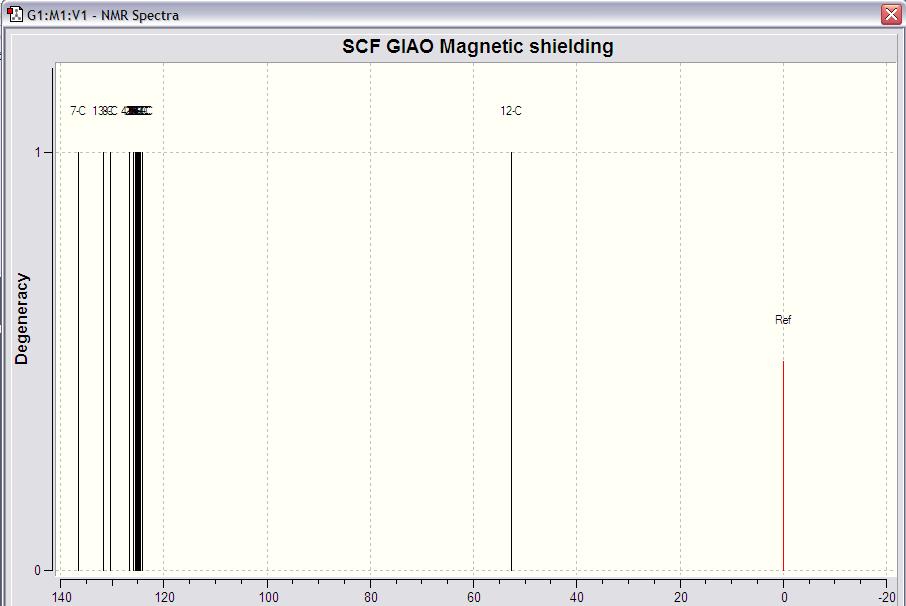
Analysis:
By comparing the 2 NMR graphs of the 2 isomers, I cannt see the difference between the 2 graphs.