Rep:Mod:fb618
NH3
NH3 Summary Information
| Molecule name | NH3 |
| Calculation method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final Energy E(RB3LYP) in au | -56.55776873 |
| RMS gradient in au | 0.00000485 |
| Point group of the molecule | C3v |
NH3 Item section
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
NH3 Jmol file
NH3 Optimised File |
The optimisation file can be viewed here
NH3 Key structural information
Optimised N-H bond distance: 1.02 Å
Optimised H-N-H bond angle: 106°
NH3 Display Vibrations
The calculation was successful as there are no negative vibrational frequencies shown in the table.
NH3 Vibrations and Frequencies
| Wavenumber in cm-1 | 1090 | 1694 | 1694 | 3461 | 3590 | 3590 |
| Symmetry label | A1 | E | E | A1 | E | E |
| IR Intensity in arbitrary units | 145 | 14 | 14 | 1 | ~ 0 | ~ 0 |
| Image |  |
 |
 |
 |
 |

|
Number of expected modes from 3N-6 rule = 3*4-6 = 6
Degeneracy
Modes with energy 1694 cm-1 and symmetry label E are degenerate. The modes with energy 3590 cm-1 and symmetry label E are also degenerate.
Bending Vibrations
The wagging mode at 1090 cm-1 is a bending vibration. It is Raman active since the polizability of the molecule changes. It is also the most IR active mode of NH3 which is reflected in the high IR intensity of 145. The mode can be IR and Raman active because NH3 does not have a centre of inversion.
The two degenerate scissoring modes at 1694 cm-1 are also bending vibrations. The modes are IR active as there is a change in dipole moment. The IR activity is reflected in the calculated IR intensity of 14. The modes are also raman active.
Bond stretches
The symmetric stretch at 3461 cm-1 is a stretching mode. It is Raman active as expected since the size of the molecule changes by a large amount, which changes the polarizablilty of the molecule. Since the stretch is symmetric though there is not change in dipole moment. Therefore the stretch is not IR active, shown by the very low IR Intensity of 1.
The two degenerate asymmetric stretches at 3590 cm-1 are also Raman active. They are not IR active because the changes in dipole moment due to the stretch cancel each other out so that the overall change in dipole moment is almost 0 as shown by the data.
Special vibrational modes
The symmetric stretch at 3461 cm-1 is also called the highly symmetric mode. It has the symmetry label A1.
Another special vibrational mode is the umbrella mode at 1090 cm-1.
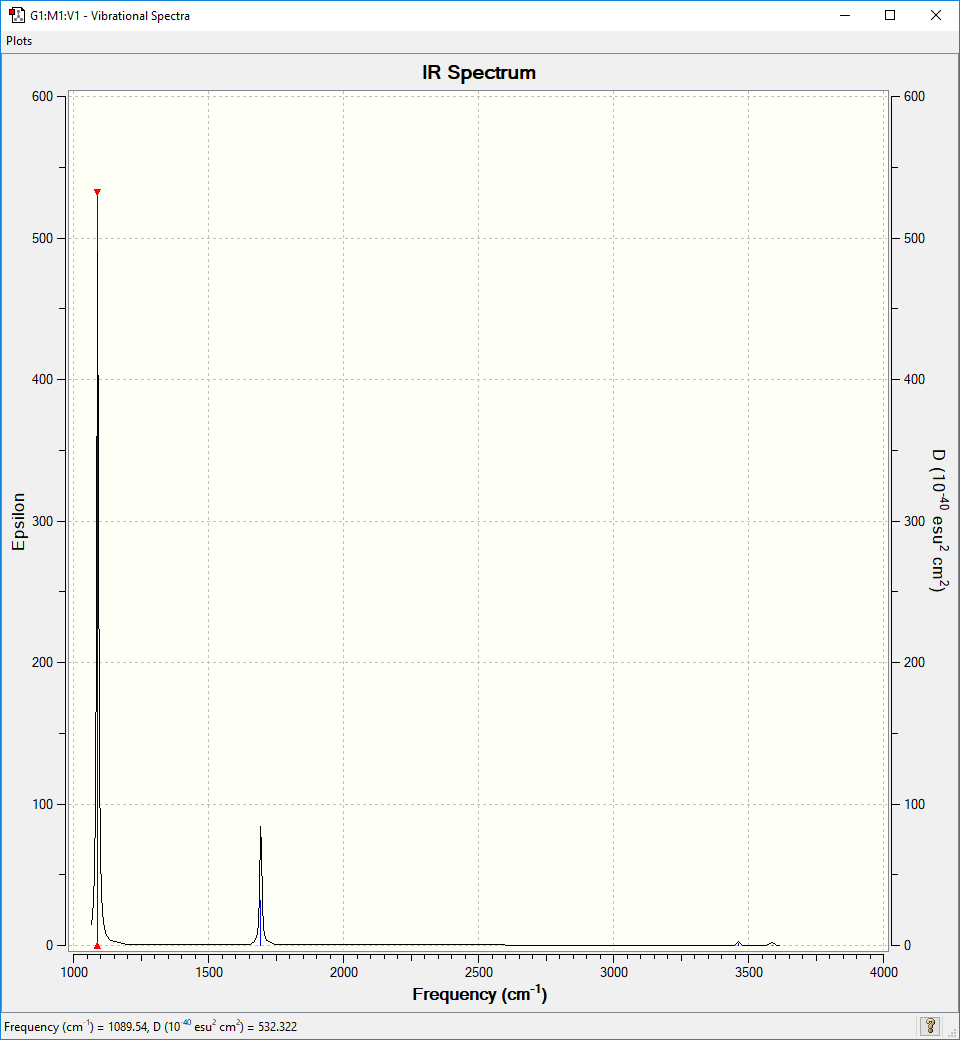
IR spectrum
2 distinct peaks are expected in the IR spectrum since there are only 2 non degenerate IR active vibrational modes with an IR intensity high enough to be observed. The peaks are expected at 1090 cm-1 and 1694 cm-1. The spectrum clearly shows these peaks.
NH3 NBO Charge Analysis
The nitrogen atom is expected to have a negative charge since it has a higher electronegativity than the hydrogen atoms.
Charge on the nitrogen atom: -1.124
Charge on the hydrogen atoms: +0.375
H2
H2 Summary Information
| Molecule name | H2 |
| Calculation method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final Energy E(RB3LYP) in atomic units | -1.17853936 |
| RMS gradient in atomic units | 0.00000017 |
| Point group of the molecule | D∞h |
H2 Item section
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES
H2 Jmol file
H2 Optimised File |
The optimisation file can be viewed here
H2 Key structural information
Optimised H-H bond distance: 0.74Å
H2 Display Vibrations
The calculation was successful as there are no negative vibrational frequencies shown in the table.
H2 Vibrations and Frequencies
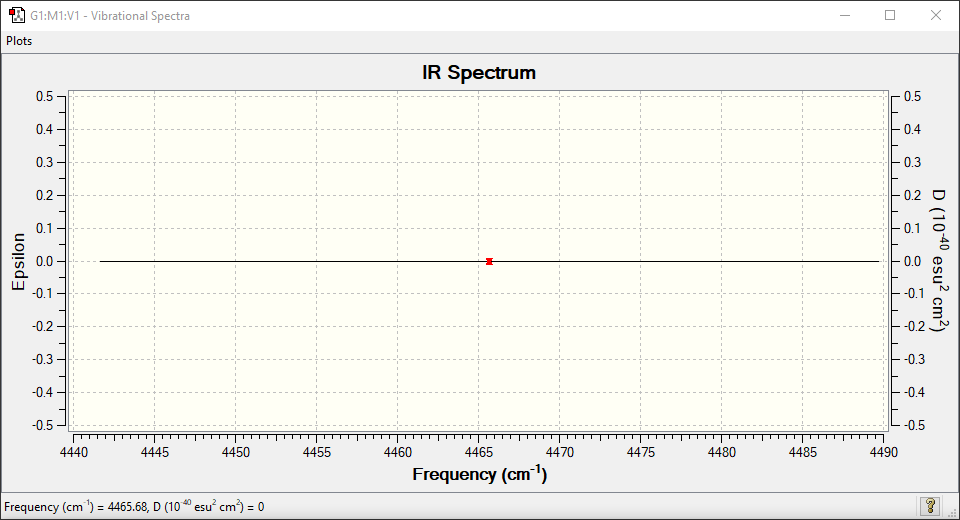
| Wavenumber in cm-1 | 4466 |
| Symmetry | SGG |
| IR Intensity in arbitrary units | 0 |
| Image | 
|
Number of expected modes from 3N-5 rule = 3*2-5 = 1
0 bands are expected in the IR spectrum since there are no IR active vibrational modes. The spectrum clearly shows a perfect flat line since there is absolutely no change in dipole moment.
H2 NBO Charge Analysis
Both hydrogen atoms are expected to have a charge of 0 because there is no electronegativity difference.
Charge on each hydrogen atom: 0
N2
N2 Summary Information
| Molecule name | N2 |
| Calculation method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final Energy E(RB3LYP) in atomic units | -109.52412868 |
| RMS gradient in atomic units | 0.00000217 |
| Point group of the molecule | D∞h |
N2 Item section
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000001 0.001800 YES
RMS Displacement 0.000002 0.001200 YES
N2 Jmol file
N2 Optimised File |
The optimisation file can be viewed here
N2 Key structural information
Optimised N-N bond distance: 1.11 Å
N2 Display Vibrations
The calculation was successful as there are no negative vibrational frequencies shown in the table.
N2 Vibrations and Frequencies
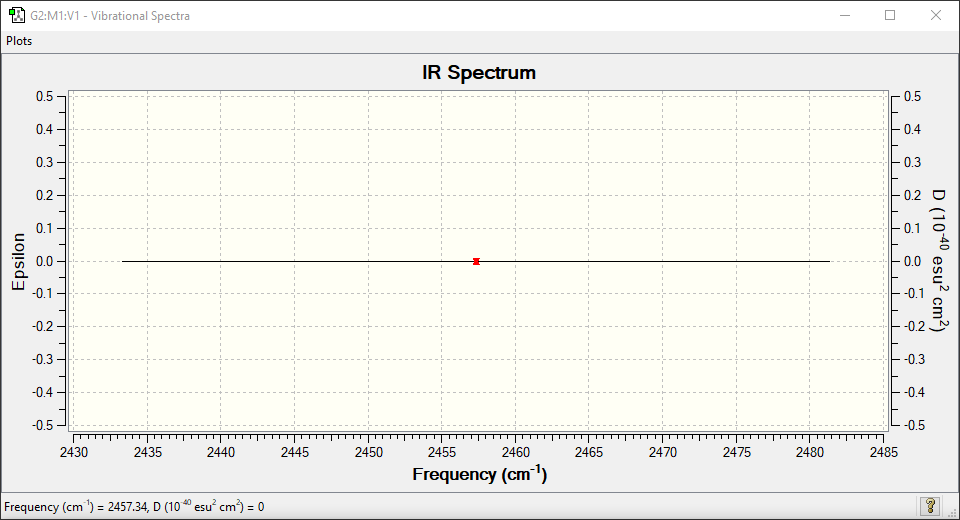
| Wavenumber in cm-1 | 2457 |
| Symmetry | SGG |
| IR Intensity in arbitrary units | 0 |
| Image | 
|
Number of expected modes from 3N-5 rule = 3*2-5 = 1
There are no peaks expected in the IR spectrum since there are no IR active vibrational modes. The spectrum below clearly shows a perfect flat line.
N2 NBO Charge Analysis
Both nitrogen atoms are expected to have a charge of 0 because there is no electronegativity difference.
Charge on each nitrogen atom: 0
N2 HOMO/LUMO
| Orbital | Image | Energy in au |
| HOMO | 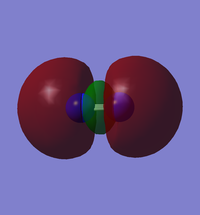 |
-0.42688 |
| LUMO |  |
-0.02412 |
N2 as ligands
| CCDC identifier of Complex with N2 ligand | DEKFUX |
| N2 bond length in complex | 1.086(6) Å |
| N2 bond length obtained | 1.11 Å |
| Link to sample structure | [[1]] |
The bond length found in the complex is shorter than the bond length obtained from the calculation. This result would mean that electron density is donated from the complex towards the ligand. This is called backdonation. However, since Ruthenium complexes are not usually known to undergo this, a computational error is more likely. Further error could be introduced through the experimental determination of the bond length.
Energies in the Haber Process
| E(NH3) in atomic units to 7 d.p. | -56.5577687 |
| 2*E(NH3) in atomic units to 7 d.p. | -113.1155375 |
| E(N2) in atomic units to 7 d.p. | -109.5241287 |
| E(H2) in atomic units to 7 d.p. | -1.1785394 |
| 3*E(H2) in atomic units to 7 d.p. | -3.5356181 |
| ΔE in atomic units to 7 d.p. | -0.0557907 |
| ΔE in kJmol-1 to 1 d.p. | -146.5 |
Ammonia gas is more stable than hydrogen gas and nitrogen gas because the overall enthalpy ΔE change is -146.5 kJmol-1 is negative.
SiH4
SiH4 Summary Information
| Molecule name | SiH4 |
| Calculation method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final Energy E(RB3LYP) in atomic units | -291.88802760 |
| RMS gradient in atomic units | 0.00000002 |
| Point group of the molecule | Td |
SiH4 Item section
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
SiH4 Jmol file
SiH4 Optimised File |
The optimisation file can be viewed here
SiH4 Key structural information
Optimised Si-H bond distance: 1.48 Å
Optimised H-Si-H bond angle: 109.47°
SiH4 Display Vibrations

The calculation was successful as there are no negative vibrational frequencies shown in the table.
SiH4 Vibrations and Frequencies
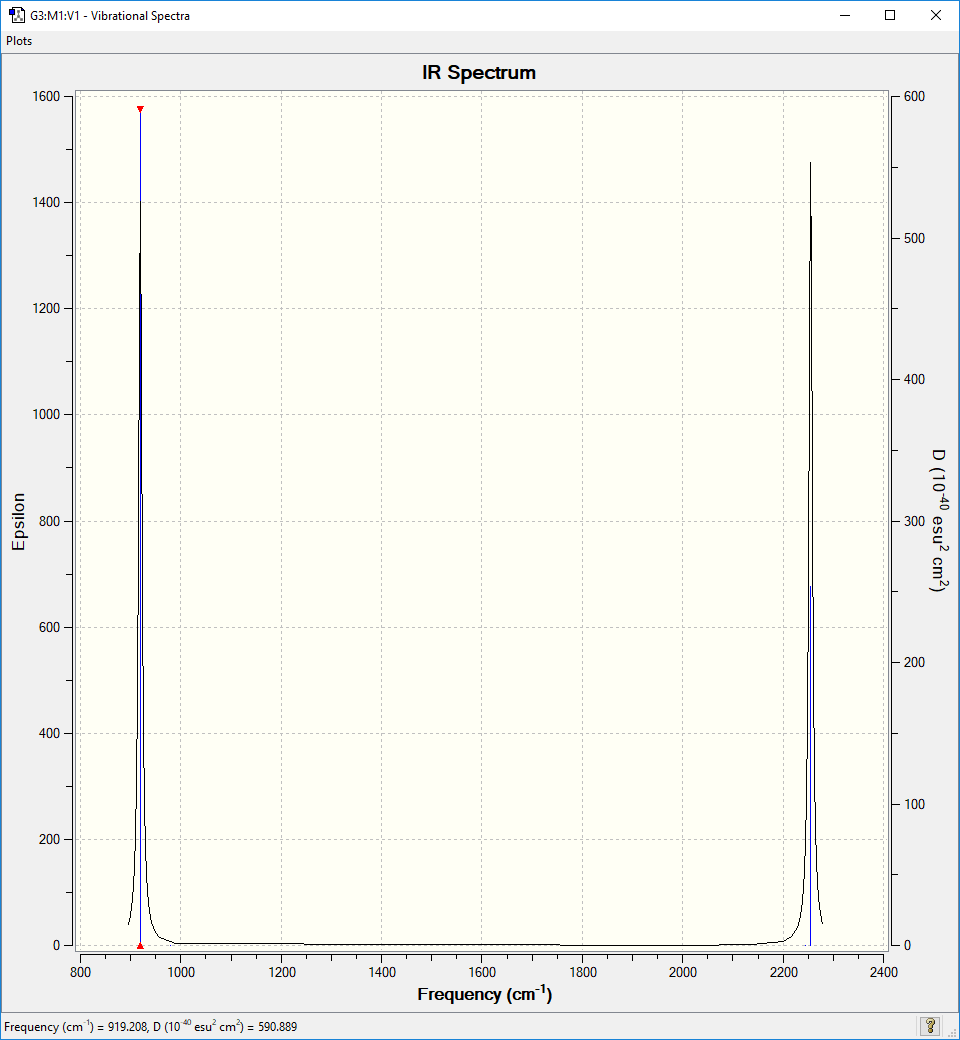
| Wavenumber in cm-1 | 919 | 919 | 919 | 979 | 979 | 2244 | 2255 | 2255 | 2255 |
| Symmetry | T2 | T2 | T2 | E | E | A1 | T2 | T2 | T2 |
| IR Intensity in arbitrary units | 136 | 136 | 136 | 0 | 0 | 0 | 143 | 143 | 143 |
| Image |  |
 |
 |
 |
 |
 |
 |
 |
|
Number of expected modes from 3N-6 rule = 3*5-6 = 9
Degeneracy
The 3 modes at 919 cm-1 and symmetry label T2 are degenerate as well as the 2 modes at 979 cm-1 and symmetry label E. The 3 modes at 2255 cm-1 and symmetry label T2 are also degenerate.
Bending Vibrations
The three degenerate wagging modes with symmetry label T2 at 919 cm-1 are bending vibrations. They are IR active as there is a change in dipole moment. They are also Raman active. The modes can be Raman active and IR active since SiH4 doesn't have a centre of inversion. The IR activity is reflected in the high IR intensity value of 136 obtained through the calculation.
The two degenerate scissoring modes with symmetry label E at 979 cm-1 are also bending vibrations. They are not IR active since there is no change in dipole moment. They are Raman active.
Bond stretches
The symmetric stretch with symmetry label A1 at 2244 cm-1 is a stretching mode. It is not IR active since all changes in Dipole moment resulting from changes in individual Si-H bond lengths cancel each other out. The mode is Raman active.
The 3 degenerate asymmetric stretches with symmetry label T2 at 2255 cm-1 are also stretching vibrations. They are the most IR active modes in the molecule since they have an IR intensity value of 143, the highest of all stretches in the molecule. The 3 modes are also Raman active.
IR spectrum
There are 2 peaks expected in the IR spectrum since there are only 2 non degenerate IR active vibrational modes. The peaks are expected at 919 cm-1 and 2255 cm-1. The spectrum clearly shows these peaks.
SiH4 NBO Charge Analysis
The silicon atom is expected to have a positive charge since it has a lower electronegativity than the hydrogen atoms.
Charge on the silicon atom: +0.629
Charge on the hydrogen atoms: -0.157
SiH4 MO analysis
Silicon electron configuration: 1s22s22p63s23p2
Hydrogen electron configuration: 1s1
1 2 3 4 5
(A1)--O (A1)--O (T2)--O (T2)--O (T2)--O
Eigenvalues -- -66.12596 -5.28056 -3.63858 -3.63858 -3.63858
1 1 Si 1S 0.99660 -0.26806 0.00000 0.00000 0.00000
2 2S 0.01292 1.02338 0.00000 0.00000 0.00000
3 2PX 0.00000 0.00000 0.00000 0.00000 0.99148
4 2PY 0.00000 0.00000 0.99148 0.00000 0.00000
5 2PZ 0.00000 0.00000 0.00000 0.99148 0.00000
6 3S -0.02764 0.07827 0.00000 0.00000 0.00000
7 3PX 0.00000 0.00000 0.00000 0.00000 0.03151
8 3PY 0.00000 0.00000 0.03151 0.00000 0.00000
9 3PZ 0.00000 0.00000 0.00000 0.03151 0.00000
10 4S 0.00432 -0.01660 0.00000 0.00000 0.00000
11 4PX 0.00000 0.00000 0.00000 0.00000 -0.00789
12 4PY 0.00000 0.00000 -0.00789 0.00000 0.00000
13 4PZ 0.00000 0.00000 0.00000 -0.00789 0.00000
14 5XX 0.00972 -0.01932 0.00000 0.00000 0.00000
15 5YY 0.00972 -0.01932 0.00000 0.00000 0.00000
16 5ZZ 0.00972 -0.01932 0.00000 0.00000 0.00000
17 5XY 0.00000 0.00000 0.00000 0.00095 0.00000
18 5XZ 0.00000 0.00000 0.00095 0.00000 0.00000
19 5YZ 0.00000 0.00000 0.00000 0.00000 0.00095
20 2 H 1S 0.00016 -0.00068 -0.00042 -0.00042 -0.00042
21 2S 0.00002 0.00028 -0.00047 -0.00047 -0.00047
22 3PX 0.00009 -0.00022 -0.00007 -0.00007 -0.00043
23 3PY 0.00009 -0.00022 -0.00043 -0.00007 -0.00007
24 3PZ 0.00009 -0.00022 -0.00007 -0.00043 -0.00007
25 3 H 1S 0.00016 -0.00068 0.00042 -0.00042 0.00042
26 2S 0.00002 0.00028 0.00047 -0.00047 0.00047
27 3PX -0.00009 0.00022 -0.00007 0.00007 -0.00043
28 3PY -0.00009 0.00022 -0.00043 0.00007 -0.00007
29 3PZ 0.00009 -0.00022 0.00007 -0.00043 0.00007
30 4 H 1S 0.00016 -0.00068 -0.00042 0.00042 0.00042
31 2S 0.00002 0.00028 -0.00047 0.00047 0.00047
32 3PX -0.00009 0.00022 0.00007 -0.00007 -0.00043
33 3PY 0.00009 -0.00022 -0.00043 0.00007 0.00007
34 3PZ -0.00009 0.00022 0.00007 -0.00043 -0.00007
35 5 H 1S 0.00016 -0.00068 0.00042 0.00042 -0.00042
36 2S 0.00002 0.00028 0.00047 0.00047 -0.00047
37 3PX 0.00009 -0.00022 0.00007 0.00007 -0.00043
38 3PY -0.00009 0.00022 -0.00043 -0.00007 0.00007
39 3PZ -0.00009 0.00022 -0.00007 -0.00043 0.00007
6 7 8 9 10
(A1)--O (T2)--O (T2)--O (T2)--O (T2)--V
Eigenvalues -- -0.54726 -0.35184 -0.35184 -0.35184 0.05053
1 1 Si 1S 0.05925 0.00000 0.00000 0.00000 0.00000
2 2S -0.27507 0.00000 0.00000 0.00000 0.00000
3 2PX 0.00000 -0.16968 0.00000 0.00000 0.00000
4 2PY 0.00000 0.00000 0.00000 -0.16968 -0.09878
5 2PZ 0.00000 0.00000 -0.16968 0.00000 0.00000
6 3S 0.58223 0.00000 0.00000 0.00000 0.00000
7 3PX 0.00000 0.40049 0.00000 0.00000 0.00000
8 3PY 0.00000 0.00000 0.00000 0.40049 0.30912
9 3PZ 0.00000 0.00000 0.40049 0.00000 0.00000
10 4S 0.13912 0.00000 0.00000 0.00000 0.00000
11 4PX 0.00000 0.09051 0.00000 0.00000 0.00000
12 4PY 0.00000 0.00000 0.00000 0.09051 1.32295
13 4PZ 0.00000 0.00000 0.09051 0.00000 0.00000
14 5XX -0.01270 0.00000 0.00000 0.00000 0.00000
15 5YY -0.01270 0.00000 0.00000 0.00000 0.00000
16 5ZZ -0.01270 0.00000 0.00000 0.00000 0.00000
17 5XY 0.00000 0.00000 0.07455 0.00000 0.00000
18 5XZ 0.00000 0.00000 0.00000 0.07455 -0.14210
19 5YZ 0.00000 0.07455 0.00000 0.00000 0.00000
20 2 H 1S 0.12832 0.17278 0.17278 0.17278 -0.08967
21 2S 0.08859 0.19100 0.19100 0.19100 -0.65470
22 3PX -0.00647 -0.00112 -0.00463 -0.00463 -0.00561
23 3PY -0.00647 -0.00463 -0.00463 -0.00112 0.01247
24 3PZ -0.00647 -0.00463 -0.00112 -0.00463 -0.00561
25 3 H 1S 0.12832 -0.17278 0.17278 -0.17278 0.08967
26 2S 0.08859 -0.19100 0.19100 -0.19100 0.65470
27 3PX 0.00647 -0.00112 0.00463 -0.00463 -0.00561
28 3PY 0.00647 -0.00463 0.00463 -0.00112 0.01247
29 3PZ -0.00647 0.00463 -0.00112 0.00463 0.00561
30 4 H 1S 0.12832 -0.17278 -0.17278 0.17278 -0.08967
31 2S 0.08859 -0.19100 -0.19100 0.19100 -0.65470
32 3PX 0.00647 -0.00112 -0.00463 0.00463 0.00561
33 3PY -0.00647 0.00463 0.00463 -0.00112 0.01247
34 3PZ 0.00647 -0.00463 -0.00112 0.00463 0.00561
35 5 H 1S 0.12832 0.17278 -0.17278 -0.17278 0.08967
36 2S 0.08859 0.19100 -0.19100 -0.19100 0.65470
37 3PX -0.00647 -0.00112 0.00463 0.00463 0.00561
38 3PY 0.00647 0.00463 -0.00463 -0.00112 0.01247
39 3PZ 0.00647 0.00463 -0.00112 -0.00463 -0.00561
11 12 13 14 15
(T2)--V (T2)--V (A1)--V (A1)--V (T2)--V
Eigenvalues -- 0.05053 0.05053 0.12286 0.22050 0.23395
1 1 Si 1S 0.00000 0.00000 0.07481 -0.01739 0.00000
2 2S 0.00000 0.00000 -0.18524 -0.12336 0.00000
3 2PX 0.00000 -0.09878 0.00000 0.00000 -0.28916
4 2PY 0.00000 0.00000 0.00000 0.00000 0.00000
5 2PZ -0.09878 0.00000 0.00000 0.00000 0.00000
6 3S 0.00000 0.00000 1.59937 -1.06382 0.00000
7 3PX 0.00000 0.30912 0.00000 0.00000 1.29279
8 3PY 0.00000 0.00000 0.00000 0.00000 0.00000
9 3PZ 0.30912 0.00000 0.00000 0.00000 0.00000
10 4S 0.00000 0.00000 -0.09324 3.88850 0.00000
11 4PX 0.00000 1.32295 0.00000 0.00000 -0.98187
12 4PY 0.00000 0.00000 0.00000 0.00000 0.00000
13 4PZ 1.32295 0.00000 0.00000 0.00000 0.00000
14 5XX 0.00000 0.00000 -0.03699 0.05295 0.00000
15 5YY 0.00000 0.00000 -0.03699 0.05295 0.00000
16 5ZZ 0.00000 0.00000 -0.03699 0.05295 0.00000
17 5XY -0.14210 0.00000 0.00000 0.00000 0.00000
18 5XZ 0.00000 0.00000 0.00000 0.00000 0.00000
19 5YZ 0.00000 -0.14210 0.00000 0.00000 -0.00915
20 2 H 1S -0.08967 -0.08967 -0.08792 -0.14418 0.01627
21 2S -0.65470 -0.65470 -0.51186 -1.07842 -0.17337
22 3PX -0.00561 0.01247 -0.00878 0.00647 -0.00174
23 3PY -0.00561 -0.00561 -0.00878 0.00647 -0.01177
24 3PZ 0.01247 -0.00561 -0.00878 0.00647 -0.01177
25 3 H 1S -0.08967 0.08967 -0.08792 -0.14418 -0.01627
26 2S -0.65470 0.65470 -0.51186 -1.07842 0.17337
27 3PX 0.00561 0.01247 0.00878 -0.00647 -0.00174
28 3PY 0.00561 -0.00561 0.00878 -0.00647 -0.01177
29 3PZ 0.01247 0.00561 -0.00878 0.00647 0.01177
30 4 H 1S 0.08967 0.08967 -0.08792 -0.14418 -0.01627
31 2S 0.65470 0.65470 -0.51186 -1.07842 0.17337
32 3PX -0.00561 0.01247 0.00878 -0.00647 -0.00174
33 3PY 0.00561 0.00561 -0.00878 0.00647 0.01177
34 3PZ 0.01247 -0.00561 0.00878 -0.00647 -0.01177
35 5 H 1S 0.08967 -0.08967 -0.08792 -0.14418 0.01627
36 2S 0.65470 -0.65470 -0.51186 -1.07842 -0.17337
37 3PX 0.00561 0.01247 -0.00878 0.00647 -0.00174
38 3PY -0.00561 0.00561 0.00878 -0.00647 0.01177
39 3PZ 0.01247 0.00561 0.00878 -0.00647 0.01177
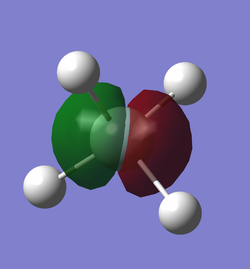
As we can see in the data table of the LOG file, for the first MO the contribution of the silicon 1s orbital is 0.99660. The contribution of the silicon 2s orbital is 0.01292. There is also some negative contribution from the silicon 3s orbital. Each hydrogen 1s orbital has a positive contribution of only 0.00016. That means it is almost a complete non bonding orbital. We can see in the diagram how small it is around the silicon atom.
The 2nd MO has a contribution of 1.02338 from the 2s orbital. There is some negative contribution from the 1s orbital and only 0.00028 positive contribution from each hydrogen 2s orbital. Therefore this orbital is also an almost complete non bonding orbital. The diagram shows that the MO is closely centred around the silicon nucleus.
The 3rd, 4th and 5th MO are degenerate and have a major contribution of 0.99148 from the silicon 2py, 2pz and 2px respectively. They also have small contributions of 0.03151 from the 3py, 3pz and 3px respectively. Each of the MOs has a slight contribution of 0.00042 from 2 of the hydrogen 1s AOs and a slight contribution of 0.00047 from 2 of the hydrogen 2s AOs. Each MO also has the corresponding negative contribution from the other 2 hydrogens. Considering the very low contributions from the hydrogen AOs, these MOs are still non bonding MOs.
The 6th lowest energy MO has a contribution of 0.58223 from the silicon 3s orbital and a contribution of 0.13912 from the 4s orbital. It also has some further very small positive and negative contributions from other silicon AOs. However it has a significant contribution of 0.12832 from each of the 4 hydrogen 1s AOs. It also has a contribution of 0.08859 from each of the hydrogen 2s AOs. This MO has significant bonding character which is represented in the diagram by the shared electron density between the hydrogen and silicon nuclei. This MO is often represented as the overlap between the in-phase hydrogen LGO with the 3s of the silicon in the same phase.
The 7th, 8th and 9th lowest energy MO are degenerate and have a contribution of 0.40049 from the silicon 3px AO, 3pz AO and 3py AO respectively. They also have a low contribution of 0.09051 from the corresponding 4p silicon AO. All three MOs have a positive contribution of 0.17278 from each hydrogen 1s AO and a contribution of 0.19100 from each hydrogen 2s AO. Each MO also has a positive contribution of 0.00463 from 4 of the 12 hydrogen 3p AOs and small negative contributions from the other hydrogen 3p AOs. These degenerate MOs are often modelled as the bonding overlap between the out-of-phase hydrogen LGOs and the silicon 3p AOs. We can see this overlap in the diagram as the 2 lobes of shared electron distribution between the silicon and 2 hydrogen atoms each. These MOs are also the highest occupied molecular orbitals, so they play essential part in the reactivity of the molecule.
Marking
Note: All grades and comments are provisional and subject to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have received your grade from blackboard.
Wiki structure and presentation 1/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES - good overall, well done.
Do you effectively use tables, figures and subheadings to communicate your work?
YES - although you could have reduced the sizes of some figures.
NH3 1/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES - excellent detailed explanations.
N2 and H2 0.5/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES - good consideration of possible factors.
Haber-Bosch reaction energy calculation 1/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 4/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
YES, overall a good explanation of the contributions from the AOs to the MOs. However please note that the contributions of the basis functions (as tabulated in the log file) do not necessarily correlations exactly with the AO contributions. Basis functions are not exact replicas of atomic AOs, and further differences will occur when molecules are modelled. I would have liked you to note that MOs 3, 4 and 5 are the bonding, in-phase MOs between Si 3p and H LGOs, and MOs 7, 8 and 9 are the corresponding antibonding MOs. In general you haven't discussed the antibonding/bonding nature of the MOs and their contributions to the bonding in the molecule. Please see the included diagram for more info.

Independence 0/1
If you have finished everything else and have spare time in the lab you could: Check one of your results against the literature, or Do an extra calculation on another small molecule, or Do some deeper analysis on your results so far