Rep:Mod:dv818
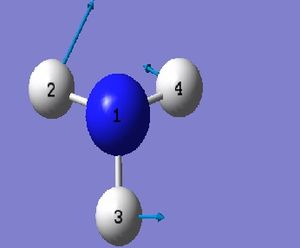
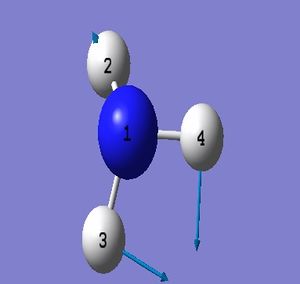
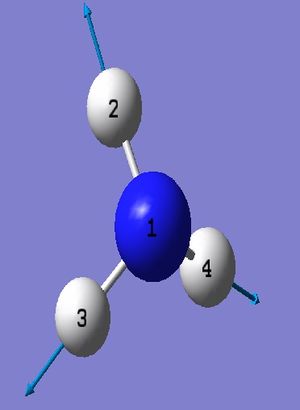
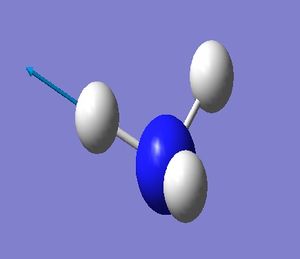
NH_3 Molecule
Calculation Method: RCAM-B3LYP
Basis Set: 6-311G(d,p)
E(RB3LYP) = -56.54485793 au
RMS Gradient Norm = 0.00022465
Point Group = C_3v
N-H Bond Length = 1.01400 Å
H-N-H Bond Angle = 107.00922 deg.
Point Group = C_3V
Item Value Threshold Converged? Maximum Force 0.000349 0.000450 YES RMS Force 0.000230 0.000300 YES Maximum Displacement 0.001447 0.001800 YES RMS Displacement 0.000644 0.001200 YES Predicted change in Energy=-5.585487D-07 Optimization completed.
NH_3 |
In 3N - 6 number of vibrational modes (nonlinear molecule), N refers to the number of atoms so 6 vibrational modes were expected (as shown by Gaussview). There are two sets of degenerate modes: 2 at frequency = 1682.61 cm**-1 and 2 at frequency = 3618.79 cm**-1). The three modes with frequency = 3498.11 and 3618.79 are stretching modes (symmetric and 2 asymmetric stretches, respectively) and the three modes with frequency = 1053.97 (additionally the umbrella mode, with H atoms moving equally forwards/backwards) cm**-1 and 1682.61 cm**-1 are bending modes (wagging and 2 scissoring, respectively) . Vertical symmetry (σ_h) is observed in A1 (1) at 1053.97 cm**-1cm**-1, horizontal symmetry (σ_v) is observed at 3498.11 cm**-1 (A1) and 3618.79 cm**-1. However, at 3498.11 cm**-1, there is further inversion symmetry (i), three perpendicular C_2 axis, and a C_3 rotation (principle axis). An IR spectrum would show 4 distinct peaks since there are two degenerate pairs of vibrations, whilst vibration at 1053.97 cm**-1 would have greatest intensity (186.4681 KM/Mole).
N (charge) = -1.020
H (charge on all) = 0.340
Since N is more electronegative than H, it is to be expected that N carries a partial negative charge and since the overall charge on the molecule must be 0 (neutral molecule), the individual charges must cancel out. The N:H charge ratio is much higher than what would be expected by simply considering the absolute (Mulliken) electronegativities of each element.
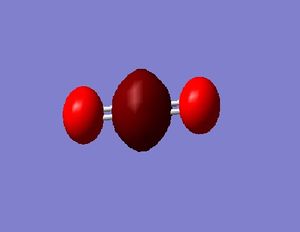
N_2 molecule:
Calculation Method: RCAM-B3LYP
Basis Set: 6-31G(d,p)
E(RB3LYP) = -109.52412868 au
RMS Gradient Norm = 0.00000282 au
N-N Bond Length = 1.10550 Å
Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES RMS Force 0.000005 0.000300 YES Maximum Displacement 0.000002 0.001800 YES RMS Displacement 0.000002 0.001200 YES Predicted change in Energy=-7.433917D-12 Optimization completed.
N_2 Optimised |
| Vibration | SGG |
|---|---|
| Frequency(cm**-1) | 2457.35 |
| IR Intensity (KM/Mole) | 0.0000 |
| Gaussview Diagram |  |
The Charge Distribution on each Nitrogen atom is 0.000, since it is a diatomic element (equal electronegativities).
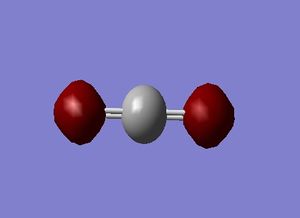
H_2 molecule
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
E(RB3LYP) = -1.17853936 au
RMS Gradient Norm = 0.00000017
Point Group : D*H
H-H Bondlength = 0.74279 Å
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES Predicted change in Energy=-1.164080D-13 Optimization completed.
H_2 Optimised |
| Vibration | SGG |
|---|---|
| Frequency(cm**-1) | 4465.68 |
| IR Intensity (KM/Mole) | 0.0000 |
| Gaussview Diagram |  |
Charge Distribution on H1 = H2 = 0.00000 since equally electronegative.
Mono-metallic Crystal Compound: DAYSUR N-N bond distance from Gaussian optimisation is 1.10550 Å whilst in DAYSUR it is 1.04 Å. Differences in Bond lengths are due to weak Coloumbic forces present in the crystal structure (a specific chemical environment). Also, presence of neighbouring delta positive Hydrogens could further stabilise the molecule, reducing the bond length.
Haber-Bosch Process Energy Analysis:
E(NH_3)= -56.54485793 au
2*E(NH_3)= -113.08971586 au
E(N_2)= -109.52412868 au
E(H_2)= -1.17853936 au
3*E(H_2)= -3.53561808 au
ΔE=2*E(NH_3)-[E(N_2)+3*E(H_2)]= -0.0299691 au ~ -78.7 kJ/Mol
Since a negative value is obtained, the forward reaction is exothermic suggesting that the product is more stable than the reactants.
CO_2 molecule:
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
E(RB3LYP) = -188.58093945
RMS Gradient Norm : 0.00000014
Point Group: D*H
C=O Bond Length = 1.16916 Å
O=C=O Bond Angle = 180.0000 deg
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES Predicted change in Energy=-8.124204D-14 Optimization completed.
CO_2 Optimised |
Charge Distribution: O1 = O2 = -0.511 , C = 1.022
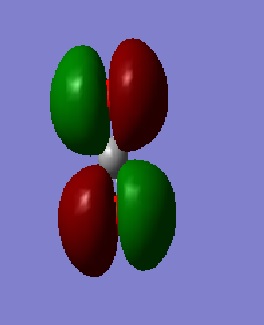
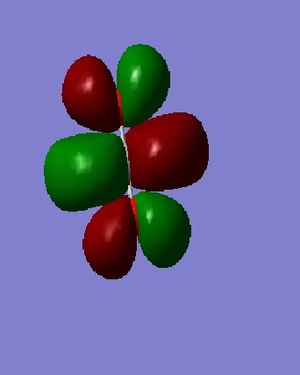
Considering MO=1 (E=-19.23659 au), the 1s Atomic Orbitals (AOs) contribute to the Molecular Orbital (MO); as such electrons are core and not valence, the overlap is small (almost negligible) as exemplified by MO=2 ,the Anti-bonding MO which is almost identical in energy. MO=3 (E= -10.38530 au) shows the sigma framework within CO_2 whilst MO=4 (E= -1.16098 au) shows the pi and sigma overlap (double bond) leading to a higher energy. M=11 (E= -0.36997 au) is the HOMO and an orbital resulting from mixing. M=12 (E = 0.02992 au) is the LUMO, thus has positive energy as there are no valence electrons that experience Coulomb forces of attraction.
Additional O_2 Molecule:
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
E(RB3LYP) = -150.25742434 au
RMS Gradient Norm = 0.00007502
Point Group: D*H
O=O Bond Length = 1.21602 Å
Item Value Threshold Converged?
Maximum Force 0.000130 0.000450 YES RMS Force 0.000130 0.000300 YES Maximum Displacement 0.000080 0.001800 YES RMS Displacement 0.000113 0.001200 YES Predicted change in Energy=-1.033738D-08 Optimization completed.
O_2 Optimised |
| Vibration | SGG |
|---|---|
| Frequency(cm**-1) | 1642.74 |
| IR Intensity (KM/Mole) | 0.0000 |
| Gaussview Diagram |  |
Charge Distribution: O1=O2= 0.0000, the atoms are identical and have identical electronegativities so there is no difference in partial charge.
Marking
Note: All grades and comments are provisional and subjecct to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have recieved your grade from blackboard.
Wiki structure and presentation 0.5/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES - however, you could have used subsections more efficiently. It is not always clear where a new section starts.
Do you effectively use tables, figures and subheadings to communicate your work?
YES
NH3 0/1
Have you completed the calculation and given a link to the file?
NO - You missed to include a link to the .log file of your finished calculation. This reduces the achievable mark for this section by 1.
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES - You correctly stated that there are two sets of degenerate modes - this explains a spectrum with 4 peaks. However there are only 2 peaks visible as peaks 4, 5 and 6 are of too low an intensity to be visible.
N2 and H2 0/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
NO - You missed to include a link to the .log file of your finished calculation. This reduces the achievable mark for this section by 1.
Crystal structure comparison 0/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
NO
Have you compared your optimised bond distance to the crystal structure bond distance?
NO
Haber-Bosch reaction energy calculation 0.5/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
NO - the energy stated for NH3 is wrong by 0.01 a.u which results in a total error of the final value. However, the sign of the energy stated is still correct.
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 2/5
Have you completed the calculation and included all relevant information?
No - You missed to include a link to the .log file of your finished calculation. This reduces the achievable mark for this section by 1.
Have you added information about MOs and charges on atoms?
You should have analysed the calculated vibrations and charge distributions as you did for NH3. You correctly stated the energies of the MOs! However, you missed to state which orbital is occupied/non-occupied. For the two lowest energy MOs you correctly identified the contributing AOs and realised that they are not significantly contributing to the bonding. However you stated them as bonding and anti-bonding in the table of displayed MOs. Actually they should be labelled as non-bonding. The same is true for MO3. This is the 1s of carbon. MO4 has no contribution from a p-orbital! it is the overlap of 2s orbitals of O and C! You correctly identified the HOMO and LUMO but missed to explain the AO contributions from MO. 6 to the LUMO and their influence on bonding.
Independence 0/1
If you have finished everything else and have spare time in the lab you could: Check one of your results against the literature, or Do an extra calculation on another small molecule, or
Yes - However, you missed to include a link to the .log file of your finished calculation. This reduces the achievable mark for this section by 1.
Do some deeper analysis on your results so far