Rep:Mod:dutchx22
NH3
Summary table
| calculation method | RB3LYP |
| Basic set | 6-31G(d.p) |
| Find energy | -56.55776873au |
| Point Group | C3V |
| Bond length | 1.01798Å |
| Bond angle | 105.741° |
Item table
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986298D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7412 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7412 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7412 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8571 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
3d jmol file
Optimised NH3 molecule |
Vibrations
Display vibrations
Wavenumber and Intensity
| wavenumber(cm-1) | 1090 | 1694 |
| symmetry | A1 | E |
| Intensity(arbitrary units) | 145 | 14 |
Answers to questions on vibrations
From the 3N-6 rule ammonia has 6 vibrational modes when you sub in N as 4. There are 2 degenerate bends at 1693.95cm-1 and 2 degenerate stretches at 3589.82cm-1. Modes that are bending are at 1089.54cm-1 and 1693.95cm-1. Modes that are stretching are at 3589.82cm-1. Bending modes are of lower energy than stretching. Bending at 1089.54cm-1, and stretching at 3461.29cm-1 are both symmetrical. Bend at 1089.54cm-1 is the umbrella mode. The number of bands expected will be 2 bands, one at 1693.95cm-1. one at 3589.82cm-1. As the other vibrational modes were symmetrical, there is no dipole formed and therefore will not be picked up on the IR spectra.
Charge distribution image
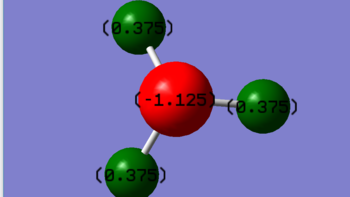
H2
Summary
| calculation method | RB3LYP |
| Basic set | 6-31G(d.p) |
| Find energy | -1.17853930au |
| Point Group | D*H |
| Bond length | 0.74309Å |
| Bond Angle | 180° |
Item table
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986298D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 0.7431 -DE/DX = -0.0002 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Jmol image of Hydrogen
Optimised H2 molecule |
Vibrations
| Wavenumber(cm-1) | 4462 |
| Symmetry | SGG |
| Intensity(arbitrary units) | 0 |
Charges
N2
Summary
| calculation method | RB3LYP |
| Basic set | 6-31G(d.p) |
| Find energy | -109.52412868au |
| Point Group | D*H |
| Bond length | 1.10550Å |
| Bond angle-180° |
Item table
Item Value Threshold Converged?
Maximum Force 0.000006 0.000450 YES
RMS Force 0.000006 0.000300 YES
Maximum Displacement 0.000002 0.001800 YES
RMS Displacement 0.000003 0.001200 YES
Predicted change in Energy=-1.248809D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1055 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Jmol image
Optimised N2 molecule |
Vibrations
| Wavenumber(cm-1) | 2457 |
| Symmetry | SGG |
| Intensity(arbitrary units) | 0 |
Charges

Comparison of bond length to TM complex
https://www.ccdc.cam.ac.uk/structures/Search?Ccdcid=ZURKIG01&DatabaseToSearch=Published
The TM complex chosen is trans-bis(Dinitrogen-N)-bis(1,2-bis(diphenylphosphino)ethane)-tungsten the ref code being ZURKIG01. The N2 bond distances are both 1.13 angstrom.This is slightly longer than the 1.10550 Angstroms in the optimised N2 molecule found. When carrying out the optimisation for N2, the computational technique applied allowed us to find the stable state equilibirum poisition. This was the position of the two nuclei that gave us the lowest possible energy, thus the most stable molecule. Therefore, what was calculate was where the slope for Potential Energy of the bond length equated to zero. The optimisation therefore did not consider in this case a W atom bonded to the N. Therefore, due to the difference in environment, the stable state equilibrium position will not be the same as now we have to consider one of the Nitrogens intereacting with a W atom. The two Nitrogens will not be able to be as close in proximity to eachother due to the presence of the W. If the two nitrogens were closer than the 1.13Å, the energy of the molecule will be greater, and thus the molecule will be less stable. Potential reasons for this change in energy could be the interaction between the N and W could be weakening the N2 interaction. Another insight maybe the N attatched to the N which is bonded to the W isn't able to get too close to the N because of the W being an energetic barrier.
Energy calculations for the Haber-Bosch process
| E(NH3) | -56.55776873au |
| 2*E(NH3) | -113.1155375au |
| E(N2) | -109.52412868au |
| E(H2) | -1.17853930au |
| 3*E(H2) | -3.5356179au |
| ΔE=2*E(NH3)-[E(N2)+3*E(H2)] | -.146.5kJ/mol |
CH4
Summary
| Calculation method | RB3LYP |
| Basis set | 6-31G(d.p) |
| Find energy | -40.52401405au |
| Point Group | TD |
| C-H bond distance | 1.09194Å |
| H-C-H bond angle | 109.471° |
Items table
Item Value Threshold Converged?
Maximum Force 0.000044 0.000450 YES
RMS Force 0.000023 0.000300 YES
Maximum Displacement 0.000124 0.001800 YES
RMS Displacement 0.000066 0.001200 YES
Predicted change in Energy=-1.089024D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.0919 -DE/DX = 0.0 !
! R2 R(1,3) 1.0919 -DE/DX = 0.0 !
! R3 R(1,4) 1.0919 -DE/DX = 0.0 !
! R4 R(1,5) 1.0919 -DE/DX = 0.0 !
! A1 A(2,1,3) 109.4712 -DE/DX = 0.0 !
! A2 A(2,1,4) 109.4712 -DE/DX = 0.0 !
! A3 A(2,1,5) 109.4712 -DE/DX = 0.0 !
! A4 A(3,1,4) 109.4712 -DE/DX = 0.0 !
! A5 A(3,1,5) 109.4712 -DE/DX = 0.0 !
! A6 A(4,1,5) 109.4712 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -120.0 -DE/DX = 0.0 !
! D2 D(2,1,5,3) 120.0 -DE/DX = 0.0 !
! D3 D(2,1,5,4) -120.0 -DE/DX = 0.0 !
! D4 D(3,1,5,4) 120.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
3d Jmol
Optimised CH4 molecule |
Vibrations
Display vibrations
Wavenumber and intensity
| Wavenumber(cm-1) | 3163 |
| Symmetry | T2 |
| Intensity(arbitrary units) | 25 |
Charges

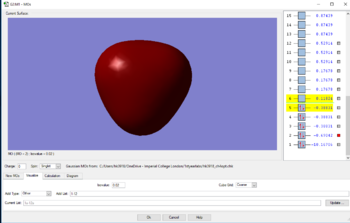
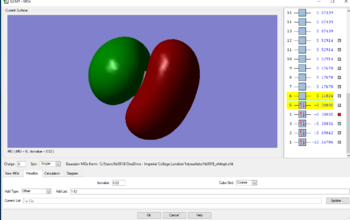
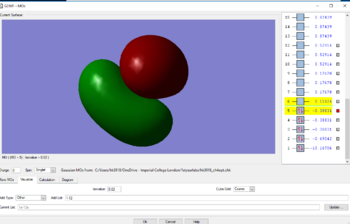
MOs for methane
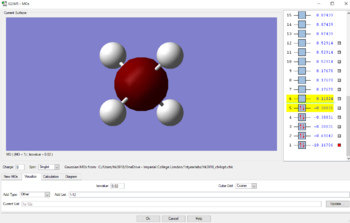
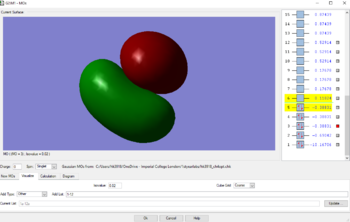
Displayed below are the 5 occupied MO of Methane. The MOs above are also all bonding. MO1 is lowest in energy, having an energy value of -10.16706au. MO1 consists of the AO of the 1s2 electron from Carbon. This is therefore depicted in the image as the electrons are held very tightly to the nuclei of the Carbon, thus allowing it to be very deep in energy.
MO2 has a higher energy of -.69042au relative to MO1. The AOs that make up MO2 are from the 2s electrons from the Carbon, and the 1s electrons from the Hydrogen. There is a good overlap as the AOs are close in energy and similar in shape, resulting in a strong interaction. The two electrons are equally distributed in MO2 between the carbon and the 4 Hydrogens as depicted in the image above.
Finally, MO3, MO4 AND MO5 are all degenerate and are in the HOMO region, with only the orientations of the MOs differing. These MOs are comprised of 2p electrons from the carbon, and 1s electrons from the H. Due to there being more nodes, the energy is slightly higher as a result of the 2p and 1s mixing.The energy as a result is -0.38831au.
Marking
Note: All grades and comments are provisional and subject to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have received your grade from blackboard.
Wiki structure and presentation 1/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES
NH3 0/1
Have you completed the calculation and given a link to the file?
NO - You missed to include a link to the .log file. This lowers the achievable mark for this section by 1.
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES - however only 2 vibrational modes are reported in this table.
Did you do the optional extra of adding images of the vibrations?
NO
Have you included answers to the questions about vibrations and charges in the lab script?
YES
N2 and H2 0/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
NO - You missed to include a link to the .log file. This lowers the achievable mark for this section by 1.
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES
Haber-Bosch reaction energy calculation 0.5/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
NO - You missed to interpret your result.
Your choice of small molecule 3.5/5
Have you completed the calculation and included all relevant information?
NO - You missed to include a link to the .log file. This lowers the achievable mark for this section by 1.
Have you added information about MOs and charges on atoms?
YES
You could have analysed the calculated vibrations as you did for NH3. You correctly commented on the occupancy, AO contributions and tried to explain the relative MO energies! Your analysis could have been a little bit more in detail, especially regarding the energy of MO2.
Independence 0/1
If you have finished everything else and have spare time in the lab you could:
Check one of your results against the literature, or
Do an extra calculation on another small molecule, or
Do some deeper analysis on your results so far
NO - No independent work has bee identified.


