Rep:Mod:dmc
The basic techniques of molecular mechanics and semi-emperical molecular orbital methods for structural and spectroscopic evaluations
The Hydrogenation of the Cyclopentadiene
Dimer 1
Dimer-1.mol |
Note: All parameters used are finalized (Quality = 4).
Iteration 36: Minimization terminated normally because the gradient norm is less than the minimum gradient norm
Stretch: 1.2923
Bend: 20.5870
Stretch-Bend: -0.8413
Torsion: 7.6715
Non-1,4 VDW: -1.4358
1,4 VDW: 4.2320
Dipole/Dipole: 0.3778
Total Energy: 31.8834 kcal/mol
Calculation completed
Dimer 2
Dimer-2.mol |
Iteration 39: Minimization terminated normally because the gradient norm is less than the minimum gradient norm
Stretch: 1.2454
Bend: 20.8603
Stretch-Bend: -0.8320
Torsion: 9.5039
Non-1,4 VDW: -1.5083
1,4 VDW: 4.3012
Dipole/Dipole: 0.4448
Total Energy: 34.0153 kcal/mol
Calculation completed
------------------------------------
The relative total energy of dimer 2 is greater than that of dimer 1. This means that dimer 2 is more stable than 1, which means that the formation of dimer 2 is the favoured reaction route, which in turn, suggests that the dimer is formed via a thermodynamic route since the relative product stability is high. This is demonstrated via the relatively high energy shown by dimer 2. For example, dimer 1 has a more positive strectching value than dimer 2. This means that the bonds in dimer 1 are weaker than that of dimer 2, since they stretch relatively further. This result agrees with the observed result of the experiment.
Product 3
Dimer-3.mol |
Iteration 39: Minimization terminated normally because the gradient norm is less than the minimum gradient norm
Stretch: 1.2067
Bend: 18.8637
Stretch-Bend: -0.7528
Torsion: 12.2396
Non-1,4 VDW: -1.5532
1,4 VDW: 5.7649
Dipole/Dipole: 0.1632
Total Energy: 35.9322 kcal/mol
Calculation completed
------------------------------------
Product 4
Dimer-4.mol |
Iteration 59: Minimization terminated normally because the gradient norm is less than the minimum gradient norm
Stretch: 1.0963
Bend: 14.5074
Stretch-Bend: -0.5493
Torsion: 12.4972
Non-1,4 VDW: -1.0507
1,4 VDW: 4.5124
Dipole/Dipole: 0.1407
Total Energy: 31.1540 kcal/mol
Calculation completed
------------------------------------
| Product | Stretching | Bending | Torsion | Van der Waals | H-bond/dipole-dipole | Total Energy |
| 3 | 1.2067 | 18.8637 | 12.2396 | 5.7649 | 0.1632 | 35.9322 |
| 4 | 1.0963 | 14.5074 | 12.4972 | 4.5124 | 0.1407 | 31.1540 |
The values for product 3 are generally higher than that of product 4. The stretching that occurs in product 3 is greater than that of product 4. This suggests that the bond lengths in 3 are longer than ‘normal’, which means that they are less stable. This also occurs for the bending, Van der Waals and H-bonding values. The torsion values, however, oppose the general trend. This might be because the double bond in product 4 is further away from the bridge than the double bond in product 3. The bridge could prevent high degrees if twisting on the double bond.
All these values show that product 4 is the more stable of the two products because its values have smaller deviations from the normal.
Stereochemistry of Nucleophilic additions to a pyridinium ring (NAD+ analogue)
Prolinol without MeMgI (5)
Prolinol-5.mol |
Iteration 169: Minimization terminated normally because the gradient norm is less than the minimum gradient norm
Stretch: 1.1986
Bend: 11.5525
Stretch-Bend: 0.0595
Torsion: 4.9284
Non-1,4 VDW: -1.9306
1,4 VDW: 11.8880
Charge/Dipole: 2.6530
Dipole/Dipole: -3.9848
Total Energy: 26.3646 kcal/mol
Calculation completed
------------------------------------
The carbonyl group, when the energy was minimised, was not the same plane as the aromatic group, which was how I drew the image originally. In fact the carbonyl group was at degrees away from the aromatic plane.
Prolinol with MeMgI (6)
Prolinol-6.mol |
Iteration 114: Minimization terminated normally because the gradient norm is less than the minimum gradient norm
Stretch: 1.4520
Bend: 14.5351
Stretch-Bend: 0.1508
Torsion: 5.2480
Non-1,4 VDW: -2.1822
1,4 VDW: 13.2282
Dipole/Dipole: -4.0705
Total Energy: 28.3614 kcal/mol
Calculation completed
------------------------------------
The aromatic ring lost it’s aromaticity during the reaction; however, the ring still remained planar. Moreover, the carbonyl group did not alter its angle away from the plane of this ring.
The methyl group attacked the molecule from the face of the molecule without the bridge. This could have been caused by steric hindrance.
Pyridinium 7 without PhNH2
Pryridinium-7.mol |
Iteration 144: Minimization terminated normally because the gradient norm is less than the minimum gradient norm
Stretch: 1.4710
Bend: 14.6508
Stretch-Bend: 0.1510
Torsion: 5.0399
Non-1,4 VDW: -2.2202
1,4 VDW: 13.2383
Dipole/Dipole: -4.0478
Total Energy: 28.2829 kcal/mol
Calculation completed
------------------------------------
This molecule had all of its aromatic rings in the same plane. The carbonyl group was at a different angle to the plane of the rings, degrees.
Pyridinium 8 with NHPh
Pryridinium-8.mol |
Iteration 258: Minimization terminated normally because the gradient norm is less than the minimum gradient norm
Stretch: 1.6427
Bend: 15.0395
Stretch-Bend: 0.2781
Torsion: -31.3965
Non-1,4 VDW: -5.9939
1,4 VDW: 14.0752
Dipole/Dipole: -5.8000
Total Energy: -12.1550 kcal/mol
Calculation completed
------------------------------------
This molecule has each aromatic ring in planes but they are at different angles to each other as opposed to the previous molecule, which had all the rings in the same plane. The NHPh phenyl ring was in the same plane as the carbonyl bond. The NHPh group attacked the molecule from the same face as the isopropyl group. This could have happened because the rings needed to twist to accommodate the attacking group, and the only way to prevent steric hindrance was to have the attacking group on the opposite side of the twisted ring.
When trying to move the oxygen of the carbonyl group to alter the starting point of optimisation, the program created this image:
The atom that moved was the nitrogen bonded to the isopropyl group. The program may have created this strange model to show that the oxygen of the carbonyl cannot be orientated in any other way because of the NHPh group; otherwise, the nitrogen would have to break off the ring.
The models could have electron withdrawing groups or electron donating groups to direct the attacking group to attack at a specific place. This would improve the model and could provide better results, for example, stretches and bends that are closer to the normal.
Stereochemistry and Reactivity of an Intermediate in the Synthesis of Taxol
Taxol 10
Taxol-10.mol |
Iteration 165: Minimization terminated normally because the gradient norm is less than the minimum gradient norm
Stretch: 2.7129
Bend: 14.5219
Stretch-Bend: 0.4049
Torsion: 20.0807
Non-1,4 VDW: -1.2177
1,4 VDW: 13.2497
Dipole/Dipole: -0.1718
Total Energy: 49.5804 kcal/mol
Calculation completed
------------------------------------
Taxol 11
Taxol-11.mol |
Iteration 367: Minimization terminated normally because the gradient norm is less than the minimum gradient norm
Stretch: 4.6434
Bend: 79.5305
Stretch-Bend: -0.0389
Torsion: 17.5543
Non-1,4 VDW: 0.1789
1,4 VDW: 25.5106
Dipole/Dipole: -0.0083
Total Energy: 127.3705 kcal/mol
Calculation completed
------------------------------------
| Isomer | Stretch | Bend | Torsion | Van der Waals | H-bonding/Dipole | Total Energy |
| 10 | 2.7129 | 14.5219 | 20.0807 | 13.2497 | -0.1718 | 49.5804 |
| 11 | 4.6434
|
79.5305 | 17.5543 | 25.5106 | -0.0083 | 127.3705 |
Generally, the values for isomer 11 are greater than that for isomer 12, except for torsion. As it was for comparison of the cyclopentadiene dimers, these results suggest that isomer 12 is the more stable of the 2 because it has values that are closer to the normal value. Furthermore, the difference in some of the values is considerable, for example the difference in the bending values and the difference in the total energy values.
How one might induce room temperature hydrolysis of a peptide
Peptide a (axial)
Peptide-a-ax.mol |
Iteration 184: Minimization terminated normally because the gradient norm is less than the minimum gradient norm
Stretch: 6.6610
Bend: 62.5242
Stretch-Bend: 0.0072
Torsion: 15.3740
Non-1,4 VDW: -1.2677
1,4 VDW: 25.7144
Dipole/Dipole: -7.4445
Total Energy: 101.5686 kcal/mol
Calculation completed
------------------------------------
Peptide a (equatorial)
Peptide-a-eq.mol |
Iteration 160: Minimization terminated normally because the gradient norm is less than the minimum gradient norm
Stretch: 6.3182
Bend: 56.1487
Stretch-Bend: -0.3240
Torsion: 13.0682
Non-1,4 VDW: -4.6705
1,4 VDW: 23.8060
Dipole/Dipole: -5.4897
Total Energy: 88.8570 kcal/mol
Calculation completed
------------------------------------
Peptide b (axial)
Peptide-b-ax.mol |
Iteration 191: Minimization terminated normally because the gradient norm is less than the minimum gradient norm
Stretch: 1.5451
Bend: 5.4044
Stretch-Bend: 0.5579
Torsion: 9.4197
Non-1,4 VDW: -4.9821
1,4 VDW: 9.4093
Dipole/Dipole: -4.5366
Total Energy: 16.8179 kcal/mol
Calculation completed
------------------------------------
Peptide b (equatorial)
Peptide-b-eq.mol |
Iteration 140: Minimization terminated normally because the gradient norm is less than the minimum gradient norm
Stretch: 1.6688
Bend: 3.7059
Stretch-Bend: 0.5242
Torsion: 7.2094
Non-1,4 VDW: -6.1675
1,4 VDW: 9.7310
Dipole/Dipole: -4.8942
Total Energy: 11.7775 kcal/mol
Calculation completed
------------------------------------
Table
| Peptide (orientation) | Total Energy |
| Peptide a (axial) | 101.5686 |
| Peptide a (equatorial) | 88.8570 |
| Peptide b (axial) | 16.8179 |
| Peptide b (equatorial) | 11.7775 |
I have found that when the models are run through the molecular mechanics, the first peptide (peptide a) is much more unstable than peptide b and it also has the amide group closer to the alcohol group than peptide b does. It has also been found that the when the N-substituent is in the axial configuration, the relative total energy of the molecule is larger, and hence, the molecule is more unstable. This instability suggests that peptide a can achieve self hydrolysis much faster than peptide b can.
These peptides can hydrolyse easily because there is free rotation around the C-N bond of the amide, thus, extreme conditions are not required for the peptide to self hydrolyse.
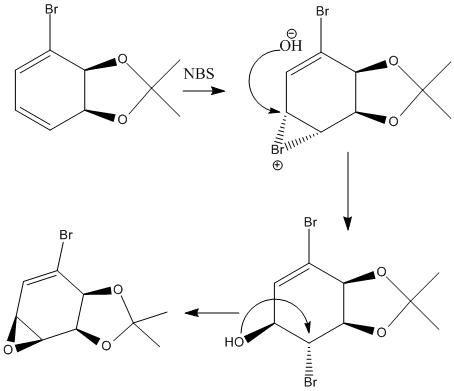
Regioselective Addition of Dichlorocarbene
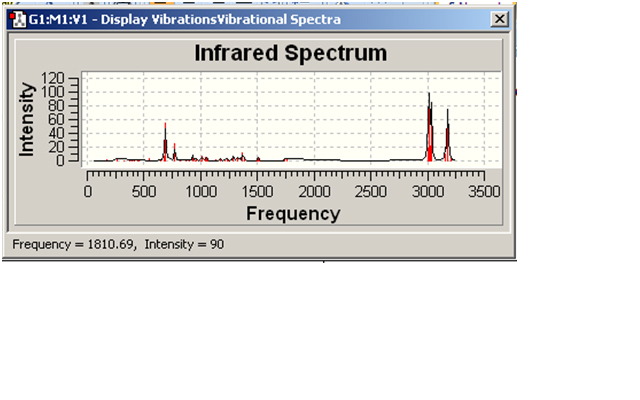
Compound 12
Compound_12_vib.mol |
| Bond type and vibration | IR peak (cm-1) |
| C-Cl stretch | 772.613 |
| C-Cl bend | 690.231 |
| C=C stretch | - |
There should be peaks in the range of 1680-1630cm-1 for the C=C stretches; however, the spectrum doesn’t show this. The strong peaks between 3000 and 3250cm-1 show the CH3 and CH2 peaks.
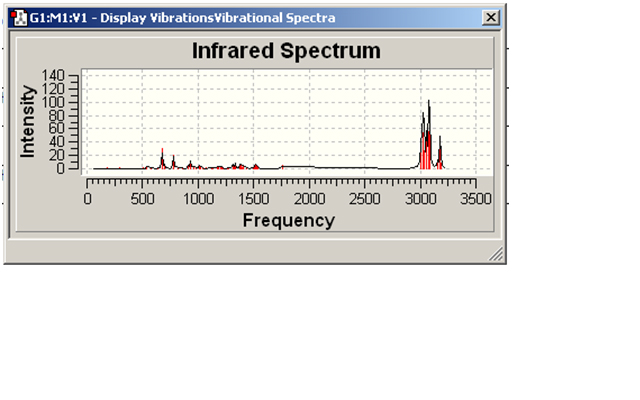
Mono-substituted compound 12
Compound_12_mono_vib.mol |
| Bond type and vibration | IR peak (cm-1) |
| C-Cl stretch | 776.971 |
| C-Cl bend | 675.068 |
| C=C stretch | - |
Again, there was no suggestion that the spectrum had any C=C peaks. This would either mean a problem with the molecules (however, the molecules show that there are C=C bonds in them) or a problem with the Gaussian Input file or the SCAN portal.
Stereoselective dissolving metal reductions
Cyclic ketone
Cyclic_ketone.mol |
Iteration 83: Minimization terminated normally because the gradient norm is less than the minimum gradient norm
Stretch: 1.9890
Bend: 3.4559
Stretch-Bend: 0.4265
Torsion: 11.6212
Non-1,4 VDW: -2.1690
1,4 VDW: 12.4113
Dipole/Dipole: 0.0339
Total Energy: 27.7688 kcal/mol
Calculation completed
------------------------------------
Cyclic alcohol
Cycalc2.mol |
Iteration 207: Minimization terminated normally because the gradient norm is less than the minimum gradient norm
Stretch: 2.0958
Bend: 3.6594
Stretch-Bend: 0.4751
Torsion: 10.9640
Non-1,4 VDW: -2.9254
1,4 VDW: 13.1534
Dipole/Dipole: 0.0117
Total Energy: 27.4338 kcal/mol
Calculation completed
------------------------------------
Mechanism:
In the mechanism the radical is created due to the electron, which is formed by the lithium dissolving in ammonia creating the [Li(NH3)x]+e- species, combining with the carbon which is bonded to the oxygen. The stereochemistry of the product lies predominantly with the endo alcohol being formed. This is due to steric effects as opposed to thermodynamic or kinetic effects of the reaction. The methyl group ortho to the alcohol forces the alcohol group to end up in the endo position as the alcohol group is a bulkier substituent than a hydrogen atom.
You could tell that the reaction had worked by performing various tests to determine that the product was an alcohol, for example use of potassium dichromate which would react with and oxidise the alcohol group to the ketone, whereas, with a ketone, no reaction would proceed. The proof that the reaction had occurred would be that the orange colour of the potassium dichromate would become a green liquid, thus, if there was no colour change then the product would be 100% ketone. Furthermore, and also less destructive, would be the use of IR and NMR spectra. IR would show peaks for either alcohol (a broad peak between 3500 and 3200cm-1) or ketone (between 1760 and 1665cm-1) stretches (or both if the reaction did not proceed to completion) stretches and bends and NMR spectra of a compound with an alcohol group would show the alcohol proton (with a peak in the range of 0.5 and 6.0ppm), which would not be apparent on an NMR spectrum of a ketone compound.
Ketone NMR (Reference used: TMS mPW1PW91/6-31G(d,p) CDCl3 GIAO)
| Carbon Number | Gaussview Value (ppm) | Literature Value (ppm) |
| C-3 | 208.871 | 213.25 |
| C-11 | 146.573 | 146.28 |
| C-13 | 109.677 | 111.06 |
| C-5 | 47.7591 | 45.66 |
| C-4 | 45.2776 | 45.24 |
| C-1 | 42.98 | 41.74 |
| C-7 | 41.5095 | 38.32 |
| C-2 | 38.8442 | 38.20 |
| C-9 | 38.2009 | 36.32 |
| C-10 | 33.238 | 33.99 |
| C-6 | 31.9513 | 27.54 |
| C-8 | 25.8855 | 23.02 |
| C-12 | 23.3122 | 22.71 |
| C-15 | 18.7231 | 16.10 |
| C-14 | 13.7825 | 11.18 |
It can be seen that there is a definite correlation between the calculated (Gaussview) and experimental (Literature) values. The main discrepancies are from the first (carbon 3) and the eleventh (carbon 6) set of values, where there are differences of nearly 5ppm in each case. This could have either come about due to an incorrect conformation produced in the calculated molecule, or it could have occurred because the calculated conditions would have been exactly the same as the real conditions when the NMR was taken.
J coupling
| Hydrogen number | Janocchio Value (Hz) | Literature Value (Hz) |
| H-1a | 12.2976 | 13.6 |
| H-4 | 7.1961 | 6.6 |
| H-14 | 6.4722 | 6.6 |
| H-5 | 11.1832 | 13.0 |
| H-4 | 12.9052 | 13.0 |
| H-6b | 2.5955 | 2.2 |
| H-2a | 12.8662 | 14.6 |
Since there are differences in which hydrogen atoms show J-coupling from the literature certain degenerate hydrogens where not recorded in Janocchio, thus, only the literature values that are closest to the calculated values are listed.
Again, the values are similar but there is variation. This also could be explained by different conformation and differing conditions since these results were taken from the NMR.
Alcohol NMR (Reference used: TMS mPW1PW91/6-31G(d,p) CDCl3 GIAO) (http://hdl.handle.net/10042/to-1019)
| Carbon Number | Gaussview Value (ppm) | Literature Value (ppm) |
| C-11 | 146.935 | 147.11 |
| C-13 | 109.556 | 110.67 |
| C-3 | 73.9199 | 76.86 |
| C-5 | 42.2233 | 43.22 |
| C-1 | 41.6673 | 39.93 |
| C-7 | 41.3496 | 39.19 |
| C-4 | 40.9524 | 38.88 |
| C-9 | 38.9667 | 37.10 |
| C-10 | 33.486 | 33.78 |
| C-2 | 30.9442 | 30.92 |
| C-6 | 30.6265 | 26.05 |
| C-8 | 25.9401 | 23.08 |
| C-12 | 24.1926 | 22.82 |
| C-15 | 18.7913 | 16.68 |
| C-14 | 15.4552 | 14.87 |
Like the results for the NMR of the ketone, there are variations between the calculated and the experimental results. The only major discrepancy in the results is for carbon number 6. This carbon had a large discrepancy between its calculated and experimental values in the ketone NMR as well. This could either mean that the configuration of this molecule has not changed much with respect to this carbon atom, hence, the similar inconsistency, or it could be coincidence that the values are quite different. However, the former seems to be the more likely reason.
J-coupling
| Hydrogen number | Janocchio Value (Hz) | Literature Value (Hz) |
| H-3 | - | 26.2 |
| H-6b | 12.2887, 2.2319 | 14.0, 2.3 |
| H-1a | 11.1633, 3.0421 | 13.2, 3.7 |
| H-14 | 6.2463 | 6.5 |
A value that could match the coupling value of 26.2Hz could not be found using the Jannochio program. This could be due to an error with the molecule created on computer, i.e. it is not real, therefore, it cannot interact the same way the actual molecule would interact in an NMR machine or it was due to an anomaly recorded in the experimental data.
The alcohol NMR generally has lesser shifts for its carbons than the carbons in the ketone, however, the values are very similar showing that there is not much change in the structure of the molecule as it is reduced. The main change in the carbon shifts is that of carbon 3, which would suggest that this is the carbon with the ketone/alcohol on it. Apart from the supposed anomaly, the J-coupling values are in the same range, however, the number of values for the alcohol from the literature is much less than that of its values for the ketone.
The optical rotation was sent to the scan 3 different times (the first two times had problems with the information in the text). However, the last time was still pending after 24 hours, thus, it was not possible to retrieve this information.
References
1. Stereoselective synthesis of (–)-4-epiaxinyssamine
Leonardo Castellanosa, Carmenza Duquea, Jaime Rodríguezb and Carlos Jiménezb, Corresponding Author Contact Information, E-mail The Corresponding Author aDepartamento de Química, Universidad Nacional de Colombia, AA 14490 Bogotá, Colombia bDepartamemto de Química Fundamental, Facultad de Ciencias, Campus da Zapateira, Universidad de A Coruña, 15071 A Coruña, Spain
Received 5 October 2006; revised 1 December 2006; accepted 11 December 2006. Available online 5 January 2007.
2. Reduction of Aliphatic and Aromatic Cyclic Ketones to sec-Alcohols by Aqueous Titanium Trichloride/Ammonia System. Steric Course and Mechanistic Implications
European Journal of Organic Chemistry
Volume 2001, Issue 12, Date: June 2001, Pages: 2235-2243
Angelo Clerici, Nadia Pastori, Ombretta Porta
3. New Active Iron Based Reducing System for Carbonyl Compounds and Imines. Stereoselective Reduction of Cyclic Ketones. ChemInform Volume 37, Issue 26, Date: June 27, 2006 Yanina Moglie, Francisco Alonso, Cristian Vitale, Miguel Yus, Gabriel Radivoy
4. Microbial Asymmetric Catalysis - Enantioselective Reduction of Ketones [New Synthetic Methods (45)] Angewandte Chemie International Edition in English Volume 23, Issue 8, Date: August 1984, Pages: 570-578 Charles J. Sih, Ching-Shih Chen