Rep:Mod:dlk1234
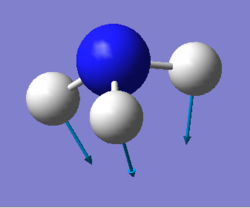
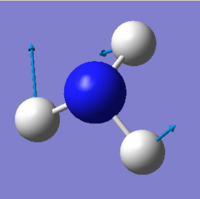
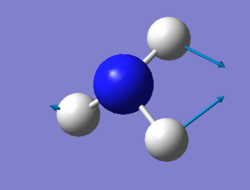
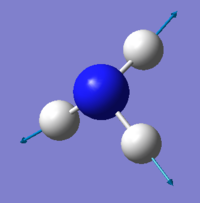
NH3 molecule
Structural Optimisation Information
Optimised NH3 molecule |
Optimised structure information: Molecule: Ammonia (NH3)
Calculation Method: RB3LYP
Basis set:6-31G(d,p)
Final energy E(RB3LYP): -56.558 a.u
RMS gradient: 0.00000485
Point group: C3v
Optimised H-N-H angle : 106°
Optimised N-H bond distance: 1.02 Å
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES
Vibrations
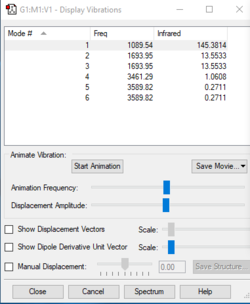
Vibrational Information:
Expected modes for ammonia: 6
Degenerate modes: 1964 and 3590 cm-1
Bending vibrations: 1090 and both 1694 cm-1
Stretching vibrations: 3461 and 3590 cm-1
Highly symmetric mode: 3461 cm-1
Umbrella mode: 1090 cm-1
Expected bands in spectrum of ammonia: 2
Charge Distribution
Charge on N atom: -1.1
Charge on H atom: 0.4
You would expect a negative charge on the N atom and a slightly positive charge on the H atom due to the difference in electronegativities, N being far more electronegative.
N2 Molecule
Structural Optimisation information
Molecule: Nitrogen (N2)
Calculation Method: RB3LYP
Basis set:6-31G(d,p)
Final energy E(RB3LYP): -109.5 a.u
RMS gradient: 0.00000011 a.u
Point group: D∞h
Optimised N-N bond distance: 1.11 Å
Optimised N-N bond angle: 180°
Optimised N2 molecule |
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES Predicted change in Energy=-1.115764D-14 Optimization completed.
Vibrations
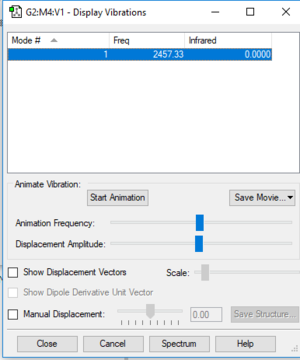
Vibrational Information:
Wavenumber: 2457.3 cm-1
Intensity arbitrary units: 0
Symmetry: SGG
Number of bands: 0 as there is no change in dipole moment, so will not appear in IR spectrum.
Charge Distribution
Charge on N atom = 0
N2 and a mono metallic TM complex
Unique Identifier : VEJSOV File:VEJSOV dlk18.mol
Optimised N2 molecule |
N-N bond length in this Transition metal complex: 1.12 Å.
N-N bond length in computational structure: 1.11 Å.
As expected, the bond N-N bond length is slightly larger when bonded to the transition metal complex because of the extra bonding to the transition metal. The extra bond being formed means the orbitals from the nitrogen atom are now interacting with the d orbital of the transition metal, lowering the electron density between the two N atoms, decreasing the bond strength and hence increasing the bond length. However the computational structure is based on theory only whereas the value from the complex is experimental, this must be taken into account especially considering the difference is only 0.01Å, which could be insignificant and due to computational error.
H2 molecule
Structural Optimisation information
Optimised H2 molecule |
Calculation Method: RB3LYP
Basis set:6-31G(d,p)
Final energy E(RB3LYP): -1.179 a.u
RMS gradient: 0.00000017 a.u
Point group: D∞h
Optimised H-H bond distance: 0.74 Å
Optimised H-H bond angle : 180°
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES Predicted change in Energy=-1.164080D-13 Optimization completed.
Vibrations
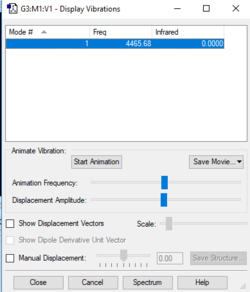
Vibrational Information:
Vibration:4465.7 cm-1
Symmetry:SGG
Intensity arbitrary units: 0
Number of bands: 0 as there is no change in dipole moment, so will not be IR active.
Charge distribution
Charge on H atom: 0
The Haber-Bosch process
N2 + 3H2 -> 2NH3
This process is used to produce Ammonia - used for fertilisers- on an industrial scale from N2 gas and H2 gas. Using the data from the optimised structures, we can calculated associated energy values for this equation.
E(NH3)= -56.55777 a.u
2*E(NH3)= -113.1154 a.u
E(N2)= -109.52413 a.u
E(H2)= -1.17854 a.u
3*E(H2)= -3.53562 a.u
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.05565 a.u
= -146.1 kJ/mol (by x 2625.5)
NF3 Molecule
Optimised NF3 molecule |
Structural Optimisation information
Molecule: Nitrogen Trifluoride (NF3)
Calculation Method: RB3LYP
Basis set:6-31G(d,p)
Final energy E(RB3LYP): -354.07 a.u
RMS gradient: 0.000068 a.u
Point group: C3v
Optimised F-N-F angle : 101.9°
Optimised N-F bond distance: 1.38 Å
Item Value Threshold Converged?
Maximum Force 0.000082 0.000450 YES
RMS Force 0.000063 0.000300 YES
Maximum Displacement 0.000356 0.001800 YES
RMS Displacement 0.000157 0.001200 YES
Predicted change in Energy=-4.598548D-08
Optimization completed.
Vibrations
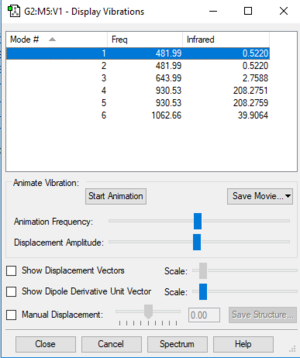
| Vibration 1 | Vibration 2 | Vibration 3 | Vibration 4 | Vibration 5 | Vibration 6 | |
|---|---|---|---|---|---|---|
| wavenumber cm-1 | 482 | 482 | 644 | 931 | 931 | 1063 |
| symmetry | E | E | A1 | E | E | A1 |
| intensity arbitrary units | 0.52 | 0.52 | 2.76 | 208.28 | 208.28 | 40.0 |
Vibrational Information:
Expected modes for NF3: 6
Degenerate modes: 482 and 931 cm-1
Bending vibrations: 482 (not clear cut, seems like one fluorine atom is stretching whilst 2 are bending) and 644 cm-1
Stretching vibrations: 1063 and 931 cm-1
Most symmetric vibration: 644 cm-1
Umbrella vibration: 644 cm-1
Number of bands in spectrum: 2
Charge Distribution
Charge on N atom: 0.66
Charge on F atom: -0.22
MO Diagrams
| MO 5 | MO 6 | MO 7 | MO 8 | MO 9 | |
|---|---|---|---|---|---|
| MO image |  |
 ] ] |
 |
 ] ] |
 ] ]
|
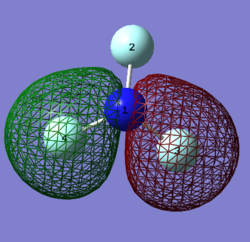
MO 5: Complete overlap of s A0s from all F atoms and N, forming bonding MOs. This particular bonding MO is the lowest energy out of all the MO images, because it has the most complete, sigma overlap.
MO 6:
The s AOs of F4 and F3 atoms are involved in this antibonding MO, this is visible as there is no overlap between the two AOs and a node is present.
MO 7:
Overlap of s AOs on F3 and F4 atom to form a bonding MO, whilst F2 s AO is antibonding.
This image is therefore a mix of antibonding and bonding MO characteristics.
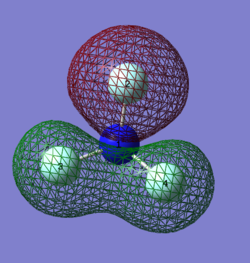
MO 8:
This image shows contribution from both s and p atomic orbitals to form this MO. Contribution of p atomic orbitals from F atoms overlapping with s atomic orbital of N to form bonding MO. Assumed it was not antibonding from all F and N s atomic orbitals because the electron density on the Nitrogen orbital is larger, suggesting p orbital contribution from F atoms.
MO 9:
This image shows a mixture of antibonding and bonding MO character, and also a contribution from both s and p atomic orbitals. F2 And N atoms' p AOs are overlapping to form bonding MO (pi), whereas F3 and F4 s AO are antibonding. This is high in energy as there is not only antibonding character,the bonding character comes from p AOs (form pi MOs) , which do not produce an as low energy overlap as s AOs (form sigma MOs)
PF5 molecule
Optimised PF5 molecule |
Calculation Method: RB3LYP
Structural optimisation summary:
Calculation Method: RB3LYP
Basis set:6-31G(d,p)
Final energy E(RB3LYP): -840.7 a.u
RMS gradient: 0.00010 a.u
Structural Information summary:
Point group: D3h
Optimised F-P-F angle : 90° and 120° in the axial and equatorial planes respectively.
Optimised P-F bond distance: 1.60 Å in axial plane and 1.57 Å in the equatorial plane.
2 angles and 2 bond lengths were found due to the Trigonal Bipyramid shape of the molecule.
Item Value Threshold Converged? Maximum Force 0.000299 0.000450 YES RMS Force 0.000090 0.000300 YES Maximum Displacement 0.000868 0.001800 YES RMS Displacement 0.000269 0.001200 YES Predicted change in Energy=-2.770927D-07 Optimization completed.
Marking
Note: All grades and comments are provisional and subjecct to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have recieved your grade from blackboard.
Wiki structure and presentation 1/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES
NH3 1/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES
N2 and H2 0/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES - however you stated a bond angle for diatomic molecules. To define a bond angle a minimum of 3 atoms is needed! You could have explained that the charges are 0 as the electronegativities are equal.
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
NO - You missed to include a links to the crystal structure. Referencing is important! However you included the unique identifier.
Have you compared your optimised bond distance to the crystal structure bond distance?
YES
Haber-Bosch reaction energy calculation 0.5/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
NO - you missed to interpret the value of the calculated reaction energy.
Your choice of small molecule 3/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
You have done a good job of presenting this information, well done! You should have explained the charges using an electronegativity argument. You cannot describe a MO as a combination of bonding and anti-bonding. Additionally you got confused about the meaning of bonding/anti-bonding and non-bonding. Just because a MO has a node it is not necessarily anti-bonding! For example MO6 and 7 are bonding MOs and not anti-bonding ones! You tried to identify the Mos contributing to the MOs. However you missed to state the principal quantum number to make your discussion easier to follow. Besides you did not get all contributing Mos right (e.g. for MO6 there is a contribution of the 2p orbital of N to the MO) Except for the MO lowest in energy you missed to comment on the relative energy and if the MOs are occupied/unoccupied.
MO7 is actually a bonding MO between the sulpher 3p and hydrogen 1s, the node in the p orbital doesn't mean it is not a bonding interaction.
Independence 1/1
If you have finished everything else and have spare time in the lab you could: Check one of your results against the literature, or Do an extra calculation on another small molecule, or
YES - well done!
Do some deeper analysis on your results so far