Rep:Mod:dl2310Module2
Introduction to Module 2: Bonding and Molecular Orbitals in Main group Compounds
Organic compounds, for the most part, demonstrate easily predictable bonding. Inorganic compounds on the other hand can behave in unexpected ways and a computational approach can be invaluable. Indeed some compounds are difficult to manipulate in a laboratory because of high toxicity or thermal instability etc.
With quantum mechanics, computational chemistry can not only be used to calculate the energy of a molecule, helping predict the most stable structures but it can also predict the location and structure of transition states that are not isolated experimentally. Thermodynamic and kinetic data can be calculated and derived as well as some properties such as dipole moments, symmetry point group, bond lengths and angles and so on.
This module aims to analyse some inorganic main group compounds through computational analysis. The software used to perform the calculations is Gaussian 09W and the results were viewed on GaussView 5.0.
The error in the energy throughout this page is ~ 10 kJ/mol, an equivalent of 0.0038088 a.u. - (1 a.u. (hartree) = 2625.50 kJ/mol).
Optimisation of the BH3, TlBr3, BBr3 Molecules
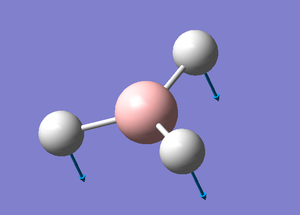
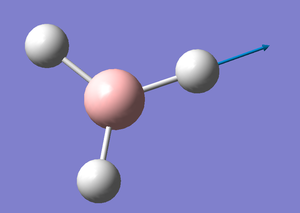
BH3
A standard trigonal planar molecule was created in GaussView and the B-H bonds were set to 1.5Å.
The B3YLP 3-21G Optimisation
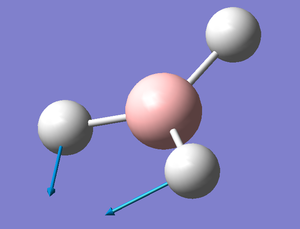
A first optimisation was then run using a B3LYP 3-21G basis set giving a better approximation of the BH3 Molecule using B3LYP 3-21G, the full log file is available here.
The inquiry button in GaussView enabled the retrieval of some data:
B-H Bond lenght: 1.19 Å
H-B-H Bond Angle: 120.0°
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 3-21G |
| Charge | 0 |
| Spin | Singlet |
| E(RB3LYP) | -26.46226338 a.u. |
| RMS Gradient Norm | 0.00020672 a.u. |
| Dipole Moment | 0.00 D |
| Point Group | D3H |
| Job Time | 12.0 seconds. |
Proof of Completion of the Calculation
Sometimes, Gaussian says that the calculations are finished when all of the optimisation parameters have not truly converged. This can be checked in the .log file. If all parameters have converged, ie the gradient tends to zero, the optimisation is complete.
Item Value Threshold Converged?
Maximum Force 0.000413 0.000450 YES
RMS Force 0.000271 0.000300 YES
Maximum Displacement 0.001610 0.001800 YES
RMS Displacement 0.001054 0.001200 YES
Predicted change in Energy=-1.071764D-06
Optimization completed.
-- Stationary point found.
The 6-31G (p,d) Optimisation
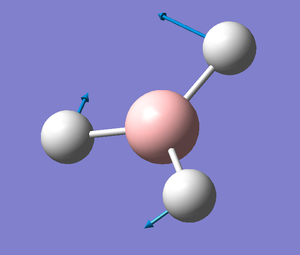
An optimisation can be improved by adding more terms to the basis set, therefore the 3-21G BH3 molecule was re-optimised using a more precise basis set, 6-31G (d,p). This calculation gave rise to this BH3 molecule optimised with the 6-31G basis set, the full .log file is available here
The inquiry button in GaussView enabled the retrieval of some data:
B-H Bond lenght: 1.19 Å
H-B-H Bond Angle: 120.0°
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G (d,p) |
| Charge | 0 |
| Spin | Singlet |
| E(RB3LYP) | -26.61532363 a.u. |
| RMS Gradient Norm | 0.00000235 a.u. |
| Dipole Moment | 0.00 D |
| Point Group | D3H |
| Job Time | 5.0 seconds. |
Proof of Completion of the Calculation
Item Value Threshold Converged?
Maximum Force 0.000433 0.000450 YES
RMS Force 0.000284 0.000300 YES
Maximum Displacement 0.001702 0.001800 YES
RMS Displacement 0.001114 0.001200 YES
Predicted change in Energy=-1.189019D-06
Optimization completed.
-- Stationary point found.
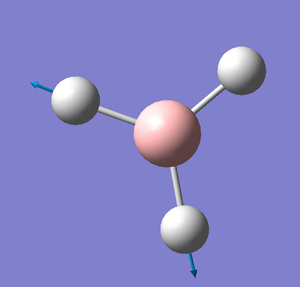
TlBr3
Pseudo-Potential Calculations
The optimisation in for this molecule is a B3YLP using a LanL2DZ basis set. It is a medium level basis set. In this model, the symmetry was manually set to D3H to get the right minimum energy. As this job demands more computational power, it was done on the SCAN HPC service and was published in D space
The inquiry button in GaussView enabled the retrieval of some data:
Tl-Br Bond length: 2.65 Å
Br-Tl-Br Bond Angle: 120.0°
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | LANL2DZ |
| Charge | 0 |
| Spin | Singlet |
| E(RB3LYP) | -91.21812851 a.u. |
| RMS Gradient Norm | 0.00000090 a.u. |
| Dipole Moment | 0.00 D |
| Point Group | D3H |
| Job Time | 37.5 seconds. |
Proof of Completion of the Calculation
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000014 0.001200 YES
Predicted change in Energy=-6.083995D-11
Optimization completed.
-- Stationary point found.
Comparison Between the Calculated Value and the Literature Value
In this section, the Tl-Br bond lenghts are compared to those found in literature to check the precision of the computational optimisation. As a molecule is never alone, the surroundings interactions play a part in the bond length that can hence vary depending on which environment the molecule is analysed in. In references, the molecules are analysed in their solid state or solution whereas the computational analysis assumes the molecule is "alone" similar to a gas state. The literature value found is 2.51Å[1]. This value is similar to the calculated value of 2.65Å. The variation can be due to the difference of states it was calculated in -as discussed above- and the approximations that are necessary to complete the calculation -basis set, quantum mechanics approximations etc. This value suggests that the computational analysis of compounds is relatively precise.
BBr3
This molecule contains a heavy atoms that need to be treated with a pseudo-potential approach and a light atom that has to be treated with a basis set. The molecule as a whole is therefore optimised with a method using a mixture of basis sets and pseudo-potentials. The 6-31G (d,p) is used for the boron atom and the LanL2DZ pseudo-potential is used fort the bromine atoms. This calculation, again, demanded hight computational power and was run on the SCAN HPC service and the results were published on D-space here
The inquiry button in GaussView enabled the retrieval of some data:
B-Br Bond length: 1.93 Å
Br-B-Br Bond Angle: 120.0°
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | Gen |
| Charge | 0 |
| Spin | Singlet |
| E(RB3LYP) | -64.43645296 a.u. |
| RMS Gradient Norm | 0.00000382 a.u. |
| Dipole Moment | 0.00 D |
| Point Group | D3H |
| Job Time | 36.0 seconds. |
Proof of Completion of the Calculation
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000036 0.001800 YES
RMS Displacement 0.000023 0.001200 YES
Predicted change in Energy=-4.026948D-10
Optimization completed.
-- Stationary point found.
Discussion
Comparison of the bond lengths for the three Molecules
| Molecule | Bond Length Å |
|---|---|
| BH3 | 1.19 |
| TlBr3 | 2.65 |
| BBr3 | 1.93 |
The length of a bond between two atoms depends on how well they can interact and that is determined by size, electronegativity, and overlap of the atomic orbitals involved in bonding. These parameters affect the strength and consequently the length of the bond formed. The stronger the interaction is, the shorter the bond. A good interaction arises from the a orbital overlap. Two atoms forming a short strong bond have orbitals of similar size and shape as well as orbitals that are not too diffuse. These parameters enable us to understand why the molecules studied have different bond lengths. Boron and Hydrogen are small atoms and have small, dense; similar sized orbitals yielding a good overlap - between the 1s Hydrogen orbital and the 1s, 2p orbitals of boron - and a strong bond. In BBr3 on the other hand, the boron center is surrounded by large electronegative atoms. The larger the atom, the more diffuse its orbitals and therefore worse orbital overlap, moreover, the electronegative nature of the bromine atoms tends to polarise the bond and induces repulsion that lengthens and weakens the bonds. Thallium is a much larger atom that has diffuse orbitals and binds weakly to the bromine atoms yielding long bonds.
Changing the ligand from hydrogen to bromine weakened the bond because of a worse overlap of atomic orbitals due to larger atom sizes and a polarisation of the bonds by electronegative bromine. Hydrogen and bromine are similar with respects to the amount of electron density they donate - one electron each- however this electron is donated from a H-s orbital and a Br-p orbital in each case giving rise to different MO diagrams, properties and reactivities. Changing the central atom also affects the bond length and strength. Boron and Thallium are in the same period of the periodic table (13) hence have similar properties, and the same number of valence electrons. As Thallium is much larger than Boron, its orbitals are extremely diffuse and produce very poor overlap with bromine giving longer bonds. In addition, both the center and the ligands are large in the thallium case which forces the atoms to sit further apart.
The Notion of Bond
The line drawn between two atoms that chemists call "bond" is subjective. In nature, there is no such thing as a clear line linking atoms but atomic orbitals overlapping between atoms. The atomic orbitals as we draw them are also subjective and only represent an area of probability to find an electron. A bond between two atoms is therefore a an area of high electron density. The bond can only happen if the overlap of the molecular orbitals and concentration of the electron density in one area leads to a lowering of the energy state of the orbitals compared to the single atom's energy levels. The way a bond is drawn as a line implies the sharing of electrons coming from both the atoms and being held on that line; however, that is a simplistic view as in reality electrons can be found much further away from the bond (from a quantum mechanics analysis). The picture is also more complicated because two atomic orbitals that overlap to form a "bond" interact and becomes a molecular orbital which has a different energy, shape and size; the electrons can therefore be found in those rather than in the original atomic orbitals. As the overlap and interaction between the atomic orbitals depends on size, energy level, and symmetry; determining where a bond will be formed and what is looks like in terms of the electron density is a very complicated affair which is why we simplify bonding by drawing a simple line. The complexity of bonding can be glimpsed from the different "types" of bonding acknowledged in the simple system; ionic, covalent, metallic each arise from a different definition of "bond" quantum mechanically.
The computational chemistry software can solve complex quantum functions and do not need the simpler "line" concept of the bond to understand it. It therefore considers a bond as a high probability of electron density and the threshold is abitrary and usually between 75-95%. GaussView does not always draw line-bonds in its structures, this does not mean that the bond is not present, it only means that the structure exceeds a pre-defined length the program recognises as "bonding length". If that happens, the atoms are too far away and the structure should be re-optimised.
Frequency and Vibrational analysis of BH3 and TlBr3
A vibrational and frequency analysis can be carried out to determine if the structure determined by the optimisation is indeed the minimum energy structure. It can be used to gives information about the reduced mass of the molecule, the symmetry of its vibrations, the change in dipole moment to be expected for each vibration etc. It also determines InfraRed spectra information for molecules.
BH3
Frequency Calculation
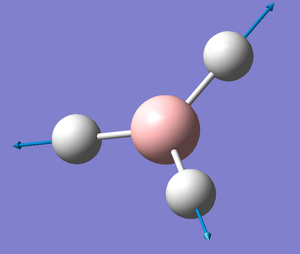
The frequency and vibrational analysis were run from the optimised structure obtained previously. It gave this structure.
B-H Bond Length: 1.19 Å
H-B-H Bond Angle: 120.0°
Summary
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final Energy | -26.61532363 a.u. |
| RMS Gradient Norm | 0.00000237 a.u. |
| Dipole Moment | 0.00 D |
| Point Group | D3H |
| Job Time | 7.0 s |
Proof of Completion
Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000002 0.000300 YES
Maximum Displacement 0.000019 0.001800 YES
RMS Displacement 0.000009 0.001200 YES
Predicted change in Energy=-1.323374D-10
Optimization completed.
-- Stationary point found.
Low frequencies --- -0.9033 -0.7343 -0.0054 6.7375 12.2491 12.2824 Low frequencies --- 1163.0003 1213.1853 1213.1880
Vibrations
The predicted IR spectrum

There are 6 vibrations calculated by Gaussian however, the IR spectrum only shows 3, this is due to the fact that two sets of vibrations are degenerate, yielding 2 peaks. One of the vibrations (4) is completely symmetrical and does not induce a change in dipole and is therefore not visible by IR spectroscopy.
TlBr3
A vibrational analysis was carried out for the pseudo-potential optimised TlBr3. The result was on D-space
Summary
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | LANL2DZ |
| Final Energy | -91.21812851 a.u. |
| RMS Gradient Norm | 0.00000088 a.u. |
| Dipole Moment | 0.00 D |
| Point Group | D3H |
| Job Time | 12.0 s |
Confirmation of competion of the job
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000011 0.001200 YES
Predicted change in Energy=-5.660901D-11
Optimization completed.
-- Stationary point found.
Low frequency information
Low frequencies --- -3.4213 -0.0026 -0.0004 0.0015 3.9367 3.9367 Low frequencies --- 46.4289 46.4292 52.1449
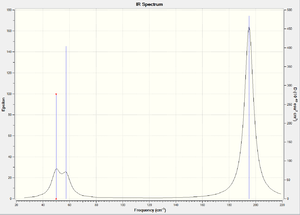
IR Spectrum
Frequency: Intensitiy
46 : 3.6 46 : 3.6 52 : 5.8 165 : 0.0 210 : 25.5 210 : 25.5
Discussion
Comparison between BH3 and TlBr3
| BH3 Frequencies | TlBr3 Frequencies | D3H symmetry label |
|---|---|---|
| 1213 | 46 | E' |
| 1213 | 46 | E' |
| 1163 | 52 | A2" |
| 2582 | 165 | A1' |
| 2715 | 210 | E' |
| 2715 | 210 | E' |
BH3
1 2 3
A2" E' E'
Frequencies -- 1163.0003 1213.1853 1213.1880
Red. masses -- 1.2531 1.1072 1.1072
Frc consts -- 0.9986 0.9601 0.9601
IR Inten -- 92.5478 14.0553 14.0589
4 5 6
A1' E' E'
Frequencies -- 2582.2624 2715.4311 2715.4323
Red. masses -- 1.0078 1.1273 1.1273
Frc consts -- 3.9595 4.8976 4.8976
IR Inten -- 0.0000 126.3307 126.3211
TlBr3
1 2 3
E' E' A2"
Frequencies -- 46.4289 46.4292 52.1449
Red. masses -- 88.4613 88.4613 117.7209
Frc consts -- 0.1124 0.1124 0.1886
IR Inten -- 3.6867 3.6867 5.8466
4 5 6
A1' E' E'
Frequencies -- 165.2685 210.6948 210.6948
Red. masses -- 78.9183 101.4032 101.4032
Frc consts -- 1.2700 2.6522 2.6522
IR Inten -- 0.0000 25.4830 25.4797
The frequency of a vibration is inversely proportional to the force constant and the reduced mass. It is observed from the tables above that the reduced mass of TlBr3 is much higher than that of BH3. The heavier atom vibrations are slower (heavier) and of lower frequency. The large difference in values for the two molecules indicate a large difference in mass (and therefore reduced mass). BH3 has stronger shorter bonds -as discussed previously- inducing higher frequencies. The ordering of the vibrations is not the same in both molecules showing a reordering of the orbitals energy. The lowest TlBr3 vibration frequency is E' symmetry whereas BH3's lowest is A2". It is observed that both E' and A2" have frequencies - and therefore energies- that are close and depending on the molecule can reorder. Both IRs show three peaks - there are 6 vibrations but only 3 appear as was discussed previously- but they differ on their epsilon values -molar extinction coefficient- as BH3 epsilon values are higher than those for TlBr3. As the molecules are similar with regards to their structures - 1 central atom surrounded by three smaller atoms in a trigonal planar arrangement - the vibrations they experience are similar, giving rise to similarly shaped IRs with different values for frequency and epsilon.
As remarked previously, the modes for the first three vibrations are fairly close in frequency and much higher up, the other three vibrations lie close together. There are two types of movement a bond can experience during a vibration, it can move but with a set length like in a pendulum case or it can be stretched and compressed like in a spring case. The two type of movements require different energies. Vibrations 1,2 and 3 -ie modes A2" and E'- do not change bond lengths and require less energy. Vibrations 4, 5 and 6 -ie modes A1' and E'- involve a change in bond lengths with demands more energy.
The energies of the same molecule optimised with different basis sets cannot be compared and as all further calculations depend on energy levels and other values, the same basis set must be used for continuity in the calculation. If studied with a different basis set, the conditions of the calculations are not the same and discrepancies in energy as big as a few hundred kJ/mol can be observed.
The Low frequencies reported after each frequency analysis represent the 3N-6 vibrations, it is the motion of the central atom. In these cases, the central atom is heavier and therefore moves more slowly with smaller amplitudes than the other atoms making those frequencies close to 0, this is a good way to check that the molecule has been optimised well and that the calculation has worked well.
Molecular Orbital Analysis
Population Analysis of BH3
The calculation results wason D-Space here
Summary
| File Type | .fch |
| Calculation Type | SP |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(D,P) |
| Total Energy | -26.61532363 a.u. |
| RMS Gradient Norm | 0.00000000 a.u. |
| Dipole Moment | 0.00 D |
| Job Time | 8.0 s |
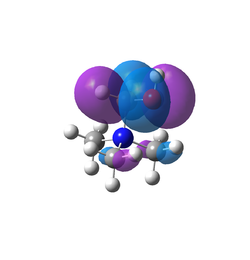
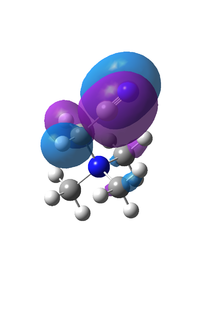
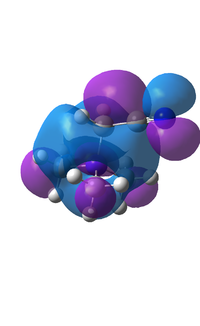
BH3 Molecular Orbital Diagram
The match between the real calculated orbitals and the LCAO orbitals are very similar. The real orbitals a broader but that is due to the fact that we only represent orbitals to about 60-70% of electron density and GaussView represents it to a higher level. The LCAO orbitals are a good representation of the electron density and of the real orbitals. This means that the LCAO approach is good enough to characterise the orbitals of small-simple molecules. The first core orbital is not represented on the diagram as it is too deep in energy to interact with the valence orbitals.
Natural Bond Orbital Analysis of NH3
Optimisation of the molecule
As the NH3 molecule is very small; the optimisation can be done directly with the higher level basis-set. The optimisation was therefore done using the RB3YLP method and the 6-31G basis set with a "nosymm" additional keyword.
NH3 Molecule using B3LYP 6-31G d,p
The full .log file can be found here.
N-H Bond Length: 1.02 Å
H-N-H Bond Angle: 105.7 °
Summary file:
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G d,p |
| Charge | 0 |
| Spin | Singlet |
| E(RB3LYP) | -56.55776856 a.u. |
| RMS Gradient Norm | 0.00000885 a.u. |
| Dipole Moment | 1.85 D |
| Point Group | C1 |
| Job Time | 14.0 seconds. |
B-H Bond Length: 1.19 Å
H-B-H Bond Angle: 120.0°
Proof of completion
Item Value Threshold Converged?
Maximum Force 0.000024 0.000450 YES
RMS Force 0.000012 0.000300 YES
Maximum Displacement 0.000079 0.001800 YES
RMS Displacement 0.000053 0.001200 YES
Predicted change in Energy=-1.629729D-09
Optimization completed.
-- Stationary point found.
Frequency Analysis
Summary
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final Energy | -56.55776856 a.u. |
| RMS Gradient Norm | 0.00000891 a.u. |
| Dipole Moment | 1.85 D |
| Point Group | C1 |
| Job Time | 10.0 s |
Confirmation of completion of the job
Item Value Threshold Converged?
Maximum Force 0.000021 0.000450 YES
RMS Force 0.000009 0.000300 YES
Maximum Displacement 0.000077 0.001800 YES
RMS Displacement 0.000039 0.001200 YES
Predicted change in Energy=-1.610513D-09
Optimization completed.
-- Stationary point found.
Low frequency information
Low frequencies --- -30.6683 -0.0014 0.0006 0.0014 20.3158 28.3272 Low frequencies --- 1089.5567 1694.1245 1694.1869
Vibration Frequencies
Frequency: Intensitiy
1089 : 145 1694 : 13.5 1694 : 13.5 3460 : 1.06 3589 : 0.27 3589 : 0.27
Population analysis for NH3
The calculation results File:NH3POPULATION ANALYSIS.LOG
Summary
| File Type | .log |
| Calculation Type | SP |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(D,P) |
| Total Energy | -56.55776856 a.u. |
| Dipole Moment | 1.85 D |
| Job Time | 4.0 s |
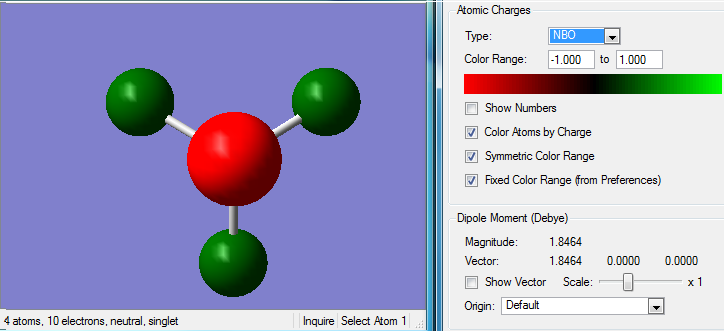
Natural Bond Orbital Analysis
Charge Distribution
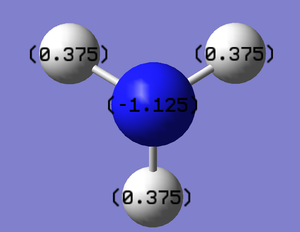
Ammonia-Borane; NH3BH3
The Ammonia-Borane molecule is larger than 4 atoms and therefore needs to be optimised at a low level basis set before being optimised at a higher level basis set to get a molecule that is well optimised.
Optimisation of the molecule using the 3-21G basis set
This optimisation gave rise to the following structure. B3LYP 3-21G Optimised Molecule The Full .log File is found here
The Following data was retrieved
B-H Bond Length : 1.21 Å N-H Bond Length : 1.02 Å B-N Bond Length : 1.67 Å H-B-H Bond Angle : 113.9° H-B-N Bond Angle : 104.5° B-N-H Bond Angle : 111.0° H-N-H Bond Angle : 107.9°
Summary
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 3-21G |
| Charge | 0 |
| Spin | Singlet |
| E(RB3LYP) | -82.76661888 a.u. |
| RMS Gradient Norm | 0.00002961 a.u. |
| Dipole Moment | 5.84 D |
| Point Group | C1 |
| Job Time | 40.0 seconds. |
The Optimisation was done with the "nosymm" keyword which explains the C1 symmetry point group displayed by the software.
Proof of completion
Item Value Threshold Converged?
Maximum Force 0.000086 0.000450 YES
RMS Force 0.000032 0.000300 YES
Maximum Displacement 0.000364 0.001800 YES
RMS Displacement 0.000192 0.001200 YES
Predicted change in Energy=-5.466107D-08
Optimization completed.
-- Stationary point found.
Optimisation of the molecule using the 6-31G d,p basis set
This optimisation gave rise to the following structure. B3LYP 6-31G d,p Optimised Molecule The Full .log File is found here
The Following data was retrieved
B-H Bond Length : 1.21 Å N-H Bond Length : 1.03 Å B-N Bond Length : 1.68 Å H-B-H Bond Angle : 113.6° H-B-N Bond Angle : 105.0° B-N-H Bond Angle : 109.6° H-N-H Bond Angle : 109.3°
Summary
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| E(RB3LYP) | -83.22469028 a.u. |
| RMS Gradient Norm | 0.00005785 a.u. |
| Dipole Moment | 5.56 D |
| Point Group | C1 |
| Job Time | 44.0 seconds. |
The Optimisation was done with the "nosymm" keyword which explains the C1 symmetry point group displayed by the software.
Proof of completion
Item Value Threshold Converged?
Maximum Force 0.000137 0.000450 YES
RMS Force 0.000038 0.000300 YES
Maximum Displacement 0.001013 0.001800 YES
RMS Displacement 0.000223 0.001200 YES
Predicted change in Energy=-1.124395D-07
Optimization completed.
-- Stationary point found.
Energy analysis
BH3: -26.61532363 a.u. NH3: -56.55776856 a.u. BH3NH3: -83.22469028 a.u.
ΔE = E(NH3BH3)-[E(NH3)+E(BH3)]
ΔE = - 0.05159809 a.u.
ΔE = - 135.47 kJ/mol
The dissociative energy of the N-B bond is therefore 135.47 kJ/mol. This is a reasonable value, a single bond is usually in the range of the few hundred kJ/mol. The literature value found for this molecule is 130 kJ/mol[2].
Mini Project: Ionic Liquids
Part 1:Comparison of selected 'onium' cations
[N(CH3)4]+
3-21G Optimisation of the Ion
This molecule is too big to be optimised immediately with a higher basis set level. It was therefore optimised using a 3-21G basis set and Published in D-Space here.
Bond lengths and angles
C-N Bond Length : 1.53Å
C-N-C Bond Angle : 109.5°
Summary
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 3-21G |
| Charge | 1 |
| Spin | Singlet |
| E(RB3LYP) | -213.01639397 a.u. |
| RMS Gradient Norm | 0.00008055 a.u. |
| Dipole Moment | 3.28 D |
| Point Group | C1 |
| Job Time | 38.9 seconds. |
Proof of Completion
Item Value Threshold Converged?
Maximum Force 0.000120 0.000450 YES
RMS Force 0.000048 0.000300 YES
Maximum Displacement 0.000503 0.001800 YES
RMS Displacement 0.000163 0.001200 YES
Predicted change in Energy=-2.930505D-07
Optimization completed.
-- Stationary point found.
6-31G d,p
The optimisation result waspublished in D-Space here
Bond lengths and angles
C-N Bond Length : 1.51Å
C-N-C Bond Angle : 109.5°
Summary
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 1 |
| Spin | Singlet |
| E(RB3LYP) | -214.18127236 a.u. |
| RMS Gradient Norm | 0.00007263 a.u. |
| Dipole Moment | 3.28 D |
| Point Group | C1 |
| Job Time | 4 minutes 26.2 seconds. |
Proof of Completion
Item Value Threshold Converged?
Maximum Force 0.000136 0.000450 YES
RMS Force 0.000051 0.000300 YES
Maximum Displacement 0.000964 0.001800 YES
RMS Displacement 0.000301 0.001200 YES
Predicted change in Energy=-3.135170D-07
Optimization completed.
-- Stationary point found.
Frequency analysis
The frequency analysis waspublished in D-Space here It gave this structure.
C-N Bond Length: 1.51 Å
C-N-C Bond Angle: 109.4°
Summary
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final Energy | -214.18127236 a.u. |
| RMS Gradient Norm | 0.00007263 a.u. |
| Dipole Moment | 3.28 D |
| Point Group | C1 |
| Job Time | 10 minutes 2.3 seconds |
Proof of Completion
Item Value Threshold Converged?
Maximum Force 0.000138 0.000450 YES
RMS Force 0.000073 0.000300 YES
Maximum Displacement 0.000821 0.001800 YES
RMS Displacement 0.000322 0.001200 YES
Predicted change in Energy=-3.044832D-07
Optimization completed.
-- Stationary point found.
Low frequencies --- -19.3355 -12.9437 -8.5264 0.0005 0.0007 0.0010 Low frequencies --- 174.3886 275.5270 282.4322
Population Analysis
The calculation results was published in D-Space here
Summary
| File Type | .chk |
| Calculation Type | SP |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(D,P) |
| Total Energy | -214.18127669 a.u. |
| RMS Gradient Norm | 0.00000000 a.u. |
| Dipole Moment | 0.00 D |
| Job Time | 15.0 s |
Molecular Orbitals
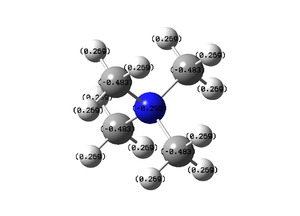
NBO, Charge Analysis
The colour range goes from red to green and from -1.7 to 1.7.
Charge on:
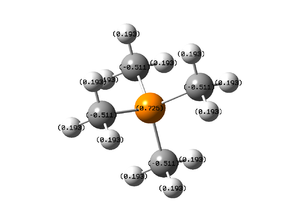
Carbon Atoms: -0.483
Nitrogen Atom: -0.295
Hydrogen Atoms: 0.269
[P(CH3)4]+
3-21G
This molecule is too big to be optimised immediately with a higher basis set level. It was therefore optimised using a 3-21G basis set and Published on D-Space here.
Bond lengths and angles
C-P Bond Length : 1.86Å
C-P-C Bond Angle : 109.5°
Summary
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 3-21G |
| Charge | 1 |
| Spin | Singlet |
| E(RB3LYP) | -498.20549670 a.u. |
| RMS Gradient Norm | 0.00002919 a.u. |
| Dipole Moment | 3.51 D |
| Point Group | C1 |
| Job Time | 2 minutes 33.2 seconds. |
Proof of Completion
Item Value Threshold Converged?
Maximum Force 0.000114 0.000450 YES
RMS Force 0.000027 0.000300 YES
Maximum Displacement 0.000683 0.001800 YES
RMS Displacement 0.000236 0.001200 YES
Predicted change in Energy=-1.274108D-07
Optimization completed.
-- Stationary point found.
6-31G d,p
The optimisation result wasPublishing on D-Space here
Bond lengths and angles
C-P Bond Length : 1.82Å
C-P-C Bond Angle : 109.4°
Summary
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 1 |
| Spin | Singlet |
| E(RB3LYP) | -500.82700338 a.u. |
| RMS Gradient Norm | 0.00001371 a.u. |
| Dipole Moment | 3.51 D |
| Point Group | C1 |
| Job Time | 5 minutes 50.5 seconds. |
Proof of Completion
Item Value Threshold Converged?
Maximum Force 0.000051 0.000450 YES
RMS Force 0.000016 0.000300 YES
Maximum Displacement 0.000925 0.001800 YES
RMS Displacement 0.000291 0.001200 YES
Predicted change in Energy=-5.202181D-08
Optimization completed.
-- Stationary point found.
Frequency Analysis
The frequency analysis wasPublished on D-Space here
It gave this structure.
C-P Bond Length: 1.82 Å
C-P-C Bond Angle: 109.5°
Summary
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final Energy | -500.82700338 a.u. |
| RMS Gradient Norm | 0.00001372 a.u. |
| Dipole Moment | 3.51 D |
| Point Group | C1 |
| Job Time | 9 minutes 47.7 seconds. |
Proof of Completion
Item Value Threshold Converged?
Maximum Force 0.000138 0.000450 YES
RMS Force 0.000073 0.000300 YES
Maximum Displacement 0.000821 0.001800 YES
RMS Displacement 0.000322 0.001200 YES
Predicted change in Energy=-3.044832D-07
Optimization completed.
-- Stationary point found.
Low frequencies --- -19.6658 -9.1081 -0.0017 -0.0010 0.0020 11.9410 Low frequencies --- 152.0274 181.9476 190.0197
Population Analysis
The calculation results was published in D-Space here
Summary
| File Type | .chk |
| Calculation Type | SP |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(D,P) |
| Total Energy | -500.82700338 a.u. |
| RMS Gradient Norm | 0.00000000 a.u. |
| Dipole Moment | 3.51 D |
| Job Time | 1 minutes 32.0 seconds. |
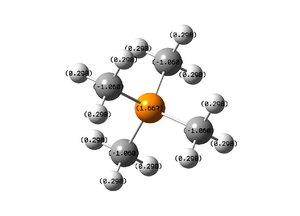
NBO, Charge Analysis
The colour range goes from red to green and from -1.7 to 1.7.
Charge on:
Carbon Atoms: -1.068
Phosphorus Atom: 1.667
Hydrogen Atoms: 0.298
[S(CH3)3]+
3-21G
This molecule is too big to be optimised immediately with a higher basis set level. It was therefore optimised using a 3-21G basis set and Published on D-Space here.
Bond lengths and angles
C-S Bond Length : 1.91Å
C-S-C Bond Angle : 100.4°
Summary
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 3-21G |
| Charge | 1 |
| Spin | Singlet |
| E(RB3LYP) | -515.05231166 a.u. |
| RMS Gradient Norm | 0.00001158 a.u. |
| Dipole Moment | 3.63 D |
| Point Group | C1 |
| Job Time | 5 minutes 46.4 seconds.. |
Proof of Completion
Item Value Threshold Converged?
Maximum Force 0.000030 0.000450 YES
RMS Force 0.000006 0.000300 YES
Maximum Displacement 0.001123 0.001800 YES
RMS Displacement 0.000455 0.001200 YES
Predicted change in Energy=-1.223280D-08
Optimization completed.
-- Stationary point found.
6-31G d,p
The optimisation result wasPublished on D-Space here
Bond lengths and angles
C-S Bond Length : 1.82Å
C-S-C Bond Angle : 102.7°
Summary
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 1 |
| Spin | Singlet |
| E(RB3LYP) | -517.68326181 a.u. |
| RMS Gradient Norm | 0.00001381 a.u. |
| Dipole Moment | 3.64 D |
| Point Group | C1 |
| Job Time | 3 minutes 53.9 seconds. |
Proof of Completion
Item Value Threshold Converged?
Maximum Force 0.000028 0.000450 YES
RMS Force 0.000013 0.000300 YES
Maximum Displacement 0.000749 0.001800 YES
RMS Displacement 0.000215 0.001200 YES
Predicted change in Energy=-2.702560D-08
Optimization completed.
-- Stationary point found.
Frequency Analysis
The frequency analysis wasPublished on D-Space here
It gave this structure.
C-S Bond Length: 1.51 Å
C-S-C Bond Angle: 109.4°
Summary
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final Energy | -517.68326181 a.u. |
| RMS Gradient Norm | 0.00001373 a.u. |
| Dipole Moment | 3.64 D |
| Point Group | C1 |
| Job Time | 4 minutes 37.9 seconds. |
Proof of Completion
Item Value Threshold Converged?
Maximum Force 0.000041 0.000450 YES
RMS Force 0.000014 0.000300 YES
Maximum Displacement 0.001094 0.001800 YES
RMS Displacement 0.000380 0.001200 YES
Predicted change in Energy=-2.743313D-08
Optimization completed.
-- Stationary point found.
Low frequencies --- -24.7798 -14.9986 -11.0843 -0.0010 0.0028 0.0041 Low frequencies --- 159.4663 192.9849 202.4751
Population Analysis
The calculation results was published in D-Space here
Summary
| File Type | .chk |
| Calculation Type | SP |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(D,P) |
| Total Energy | -517.68326181 a.u. |
| RMS Gradient Norm | 0.00000000 a.u. |
| Dipole Moment | 3.64 D |
| Job Time | 58.8 seconds. |
NBO, Charge Analysis
The colour range goes from red to green and from -1.7 to 1.7.
Charge on:
Carbon Atoms: -0.846
Sulfur Atom: 0.917
Hydrogen Atoms: 0.297
Discussion
Comparison of structures and bond lengths
| Cation | C-X Bond Length | C-X-C Bond Angle | Structure |
|---|---|---|---|
| [N(CH3)4]+ | 1.51 Å | 109.5° | Tetrahedral |
| [P(CH3)4]+ | 1.82 Å | 109.4° | Tetrahedral |
| [S(CH3)3]+ | 1.82Å | 102.7° | Distorted Trigonal Planar |
The nitrogen and phosphorus based ions adopt tetrahedral structures which minimises steric crowding ie. mutual repulsion of the methyl groups. This is confirmed by the bond angles determined by the optimisations. The nitrogen ion has a C-N-C angle of 109.5° and the phosphorus ion has a C-P-C angle of 109.4°; this is almost a perfect tetrahedral arrangement (109.7°).
As the sulphur ion only has three methyl groups surrounding the molecule, it could be expected to be trigonal planar, however, a trigonal pyramidal structure is observed instead. The sulphur atom has a large lone pair which pushes the methyl groups together to avoid repulsion and forming a tetrahedral type pseudo-structure. A perfect trigonal pyramidal structure has C-X-C angles of 107°, however, the lone pair distorts the molecule more and the angles observed are of 102.7°.
It can be noticed that the C-N bond is shorter and stronger than the equivalent C-P and C-S bonds. The longer bond for P and S are maintly due to the fact that they are second row elements and are bigger with more diffuse orbitals and therefore worse overlap. N and C are in the same row and of similar sizes with good overlap yielding stronger bonds.
NBO Comparison
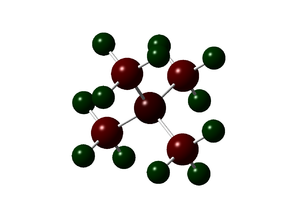 |
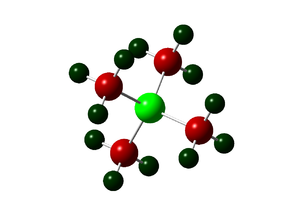 |
 |
| Cation | X= | Charge on X | Charge on C | Charge on H |
|---|---|---|---|---|
| N(CH3)4]+ | N | -0.295 | -0.483 | 0.269 |
| [P(CH3)4]+ | P | 1.667 | -1.068 | 0.298 |
| [S(CH3)3]+ | S | 0.917 | -0.846 | 0.297 |
The charge distribution can be rationalised based on electronegativity and charge of the atoms in the molecule. The Pauling electronegativity of the atoms we are working with increases thus: P < H < C < S < N. The larger the atom, the larger the effective nuclear charge (Zeff).
The phosphorus is less electronegative than carbon and carries a high positive charge with an electron donating effect onto the carbons which show a large negative charge. N is the most electronegative atom present and in the nitrogen ion it pulls electron density away from the carbon atoms and concentrates it around itself. The next electronegative atom is the sulfur and it shows similar electronegativity to carbon which means that there is a certain balance there. The S atom collects some of the electron density from the carbon atoms but the effect is much smaller than for N. The phosphorus is less electronegative than carbon and therefore carries a high positive charge with an electron donating effect onto the carbons which show a large negative charge. The charge around the hydrogens is very similar in all cases with a range of +0.269 to +0.298 which shows that they are not affected by the electronegativity of the center atom.
The charge distribution is a clear indicator of the contributions to the C-X bonds. An atom which has a high electron density held around itself contributes the most to the bond than the lower electronic density atom. The bigger the difference in electronegativity between the C and X, the more electron density X has pulled off C and the larger the difference in contributions.
| [N(CH3)4]+ | ΔχC-N=0.49 | contribution factor ~34:66 |
| [P(CH3)4]+ | ΔχC-P=0.36 | contribution factor ~60:40 |
| [S(CH3)3]+ | ΔχC-S=0.03 | contribution factor ~49:51 |
The position of the charge on [N(CH3)4]+
The nitrogen centered ion is often drwn with a localised charge on the nitrogen center. The charge distribution results calculated show that the nitrogen center is not where the positive charge, it is even slightly negatively charged. Nitrogen is electronegative and pulls electron density towards itself. The "real" picture shows that the charge is actually delocalised over the hydrogens. It is usually better and more accurate to assign the charge to the whole molecule unless a charge analysis has been done and a charge is clearly assignable to a single atom.
Part 2: Influence of functional groups
[N(CH3)3(CH2OH)]+
3-21G Optimisation
This molecule is too big to be optimised immediately with a higher basis set level. It was therefore optimised using a 3-21G basis set and Published in D-Space here.
Bond lengths and angles
C-N Bond Length : 1.54Å
O-H Bond Length : 0.99Å
C-O Bond Length : 1.42Å
C-N-C Bond Angle : 108.8°
N-C-O Bond Angle : 103.7°
Summary
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 3-21G |
| Charge | 1 |
| Spin | Singlet |
| E(RB3LYP) | -287.80633326 a.u. |
| RMS Gradient Norm | 0.00001477 a.u. |
| Dipole Moment | 4.26 D |
| Point Group | C1 |
| Job Time | 3 minutes 21.7 seconds. |
Proof of Completion
Item Value Threshold Converged?
Maximum Force 0.000036 0.000450 YES
RMS Force 0.000008 0.000300 YES
Maximum Displacement 0.000600 0.001800 YES
RMS Displacement 0.000154 0.001200 YES
Predicted change in Energy=-1.610389D-08
Optimization completed.
-- Stationary point found.
6-31G (d,p) Optimisation
The optimisation result wasPublishing on D-Space here
Bond lengths and angles
C-P Bond Length : 1.82Å
C-P-C Bond Angle : 109.4°
Summary
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 1 |
| Spin | Singlet |
| E(RB3LYP) | -289.39322212 a.u. |
| RMS Gradient Norm | 0.00001570 a.u. |
| Dipole Moment | 4.24 D |
| Point Group | C1 |
| Job Time | 13 minutes 26.4 seconds. |
Proof of Completion
Item Value Threshold Converged?
Maximum Force 0.000042 0.000450 YES
RMS Force 0.000009 0.000300 YES
Maximum Displacement 0.001218 0.001800 YES
RMS Displacement 0.000370 0.001200 YES
Predicted change in Energy=-3.630759D-08
Optimization completed.
-- Stationary point found.
Frequency Analysis
The frequency analysis waspublished on D-Space here
It gave this structure.
Summary
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final Energy | -517.68326181 a.u. |
| RMS Gradient Norm | 0.00001373 a.u. |
| Dipole Moment | 3.64 D |
| Point Group | C1 |
| Job Time | 4 minutes 37.9 seconds. |
Proof of Completion
Item Value Threshold Converged?
Maximum Force 0.000041 0.000450 YES
RMS Force 0.000014 0.000300 YES
Maximum Displacement 0.001094 0.001800 YES
RMS Displacement 0.000380 0.001200 YES
Predicted change in Energy=-2.743313D-08
Optimization completed.
-- Stationary point found.
Low frequencies --- -24.7798 -14.9986 -11.0843 -0.0010 0.0028 0.0041 Low frequencies --- 159.4663 192.9849 202.4751
MO Analysis
The calculation results was published in D-Space here
Summary
| File Type | .chk |
| Calculation Type | SP |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(D,P) |
| Total Energy | -289.39322212 a.u. |
| RMS Gradient Norm | 0.00000000 a.u. |
| Dipole Moment | 4.24 D |
| Job Time | 1 minutes 50.5 seconds. |
NBO, Charge distribution analysis
The colour range goes from red to green and from -1 to 1.
Charge on:
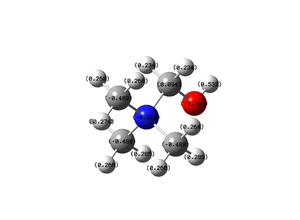
Oxygen Atom: -0.757
Hydrogen of the Hydroxyl: 0.532
Nitrogen Atom: -0.313
Carbon Atoms (CH3): -0.488
Hydrogen Atoms(CH3): 0.264 to 0.285
Hydrogen Atoms(CH2): 0.234
[N(CH3)3(CH2CN)]+
3-21G Optimisation
This molecule is too big to be optimised immediately with a higher basis set level. It was therefore optimised using a 3-21G basis set and the full .log File.
Bond lengths and angles
C-N Bond Length (CH2) : 2.75 Å
C-N Bond Length (CH3) : 1.48 Å
C-N Bond Length (CN): 1.18 Å
C-C Bond Length (C-CN): 1.38 Å
CH3-N-CH3 Bond Angle : 112.6°
CH3-N-CH2 Bond Angle : 102.0°
N-CH2-C(N) Bond Angle : 89.6°
CH2-C-N Bond Angle : 177.8°
Summary
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 3-21G |
| Charge | 1 |
| Spin | Singlet |
| E(RB3LYP) | -304.89803339 a.u. |
| RMS Gradient Norm | 0.00002208 a.u. |
| Dipole Moment | 4.14 D |
| Point Group | C1 |
| Job Time | 33 minutes 22.0 seconds. |
Proof of Completion
Item Value Threshold Converged?
Maximum Force 0.000050 0.000450 YES
RMS Force 0.000011 0.000300 YES
Maximum Displacement 0.001741 0.001800 YES
RMS Displacement 0.000686 0.001200 YES
Predicted change in Energy=-5.732318D-08
Optimization completed.
-- Stationary point found.
6-31G (d,p) Optimisation
The optimisation result was done on gaussian as the HPC got stuck and nothing ran for over 12 hours. The full.log File
Bond lengths and angles
C-N Bond Length (CH2) : 1.53Å
C-N Bond Length (CH3) : 1.51Å
C-N Bond Length (CN): 1.16 Å
C-C Bond Length (C-CN): 1.46 Å
CH3-N-CH3 Bond Angle : 109.7°
CH3-N-CH2 Bond Angle : 110.1°
N-CH2-C(N) Bond Angle : 111.7°
CH2-C-N Bond Angle : 178.9°
Summary
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 1 |
| Spin | Singlet |
| E(RB3LYP) | -306.39376664 a.u. |
| RMS Gradient Norm | 0.00002719 a.u. |
| Dipole Moment | 2.74 D |
| Point Group | C1 |
| 1 hours 55 minutes 47.0 seconds. |
Proof of Completion
Item Value Threshold Converged?
Maximum Force 0.000032 0.000450 YES
RMS Force 0.000007 0.000300 YES
Maximum Displacement 0.001679 0.001800 YES
RMS Displacement 0.000295 0.001200 YES
Predicted change in Energy=-7.709320D-08
Optimization completed.
-- Stationary point found.
Frequency Analysis
The frequency analysis waspublished on D-Space here
It gave this structure.
Summary
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final Energy | -306.39376664 a.u. |
| RMS Gradient Norm | 0.00002719 a.u. |
| Dipole Moment | 2.74 D |
| Point Group | C1 |
| Job Time | 14 minutes 51.5 seconds.. |
Proof of Completion
Item Value Threshold Converged? Maximum Force 0.000085 0.000450 YES RMS Force 0.000027 0.000300 YES Maximum Displacement 0.002677 0.001800 YES RMS Displacement 0.000820 0.001200 YES Predicted change in Energy=-2.281694D-07 Optimization completed. -- Stationary point found.
Low frequencies --- -3.9323 -0.0004 -0.0003 0.0010 5.4265 14.1052 Low frequencies --- 91.0040 153.7445 211.2101
MO Analysis
The calculation results .log file
Summary
| File Type | .chk |
| Calculation Type | SP |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(D,P) |
| Total Energy | -306.39376664 a.u. |
| RMS Gradient Norm | 0.00000000 a.u. |
| Dipole Moment | 2.74 D |
| Job Time | 3 minutes 59.0 seconds. |
NBO, Charge distribution analysis
The colour range goes from red to green and from -1 to 1.
Charge on:
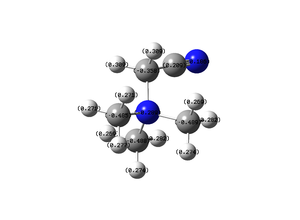
Central Nitrogen Atom: -0.289
Nitrogen of the cyano group: -0.186
Carbon of the cyano group: 0.209
Carbon of the (CH2): -0.358
Carbon Atoms (CH3): -0.485 to -0.489
Hydrogen Atoms(CH3): 0.269 to 0.282
Hydrogen Atoms(CH2): 0.309
Discussion
The effect of the OH and CN groups
 |
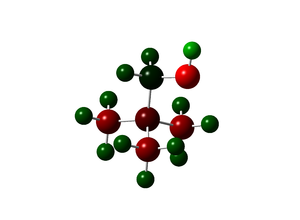 |
 |
| Functional Group | Charge on central N | Charge on functional group | Charge on C (CH3) | Charge on C (CH2) | Charge on H (CH3) | Charge on H (CH2) |
|---|---|---|---|---|---|---|
| CH3 | -0.295 | C : -0.483, H : 0.269 | -0.483 | 0.269 | ||
| OH | -0.313 | O : -0.757, H : 0.532 | -0.488 | -0.358 | 0.264 to 0.285 | 0.234 |
| CN | -0.289 | C : 0.209, N : -0.186 | -0.485 to -0.489 | -0.358 | 0.269 to 0.282 | 0.309 |
The charge distribution picture shows that OH is indeed an electron withdrawing group as the electron density is pulled away from the CH2 carbon that appears green (and not red), it is also shown by the charge numbers in the table. It also makes the hydroxyl electron even more electronegative. It is also electron donating via resonance with N and that explains that the electron density increases on the nitrogen. The charge does not change much for the three methyl groups. The CN group is electron withdrawing and everything around it has lost a bit of electron density and their charges have increased slightly.
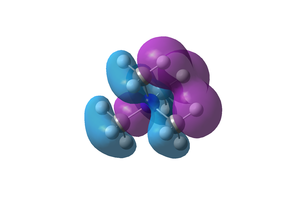
Comparison of the two compounds' HOMO and LUMO
The shape of the HOMO orbitals changes dramatically when a substituent is added. The substituted ions have localised dense orbitals that contain about half of the atomic orbitals. LUMO orbitals are more similar as the general shape is the same with addition of other orbitals on the substituent, however, this orbital is much larger and delocalised as it encompasses the whole of the molecule and its atomic orbitals. As a general rule the number of nodes and nodal planes increases with the substituents making the substituted HOMOs ans LUMOs more antibonding and less likely to be occupied. Both substituted ions have higher energy HOMOs and LUMOs but the energy gap is smaller indeed, the HOMO-LUMO energy gaps have been calculated to be 0.44609, 0.34639 and 0.31867 a.u. respectively (CH3, OH, CN). These are rather large gaps, the larger the gap is, the more difficult it is to promote an electron into the LUMO and therefore the more stable and unreactive the molecule is. It also influences the physical properties of the molecule like the melting point, the colour and magnetic properties of inorganic complexes. The substituted complexes have smaller gaps and are therefore more likely to react with incoming reactants. As both the HOMO and LUMO are antibonding, the more electron density, the more antibonding character and the higher in energy the orbital and molecule will be. This is consistent with the data as the OH group is electron donating and is higher in energy. The cyano group is slightly more reactive than the OH group as it posesses a smaller HOMO-LUMO gap. However, the overal reactivity of a molecule is not just determined by their molecular orbitals. The type of reactivity that the molecule experiences also depends on how high in energy the HOMO-LUMO couple and other factors not explored here.