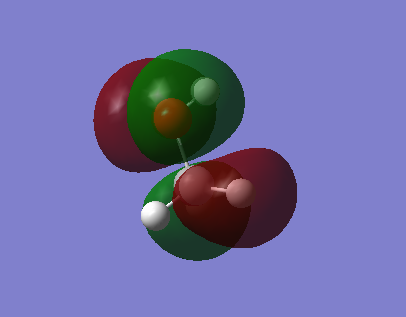Rep:Mod:dguy95
NH3 molecule
Key Information
Molecule: NH3
Calculation Method: RB3LYP
Basis Set: 6-31G(d.p)
E(RB3LYP): -56.55776873 a.u.
RMS Gradient: 0.00000485 a.u.
Point Group: C3V
N-H Bond Length: 101.798 pm
H-N-H Bond Angle: 105.741°
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986292D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7412 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7412 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7412 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8571 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
NH3 |
The optimisation file is linked to here
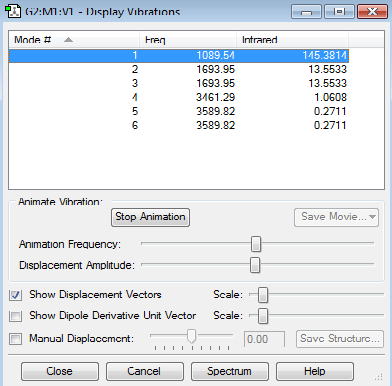
Vibrations
Expected number of modes: 6
Degenerate modesː 2,3 and 5,6
Bending Vibrationsː 1,2,3
Bond Stretchesː 4,5,6
Highly Symmetric one is 4
The Umbrella on is 1
Expected number of bands in a spectrum is 4
Charge on N = -1.125
Charge on H = 0.375
The N is expected to have a negative charge and H is expected to have a positive charge. This is because N is more electronegative than H and hence the N pulls the electron pair in the bonds towards itself
Molecular orbitals
Highest occupied molecular orbital. It is a sigma anti bonding orbital (occupied). AOs that contribute are the 1s of H and 2p of N
Lowest unoccupied molecular orbital. It is not clear what sort of MO it is and therefore which AOs are contributing.
N2
Key Information
Molecule: N2
Calculation Method: RB3LYP
Basis Set: 6-31G(d.p)
E(RB3LYP): -109.52412868 a.u.
RMS Gradient: 0.0000006 a.u.
Point Group: D̽ H
N-N Bond Length: 110.550 pm
H-N-H Bond Angle: 180°
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.400991D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1055 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
N2 |
The optimisation file is linked to here
Vibrations
No negative frequencies
H2
Key Information
Molecule: H2
Calculation Method: RB3LYP
Basis Set: 6-31G(d.p)
E(RB3LYP): -1.17853936 a.u.
RMS Gradient: 0.00000017 a.u.
Point Group: D̽ H
N-N Bond Length: 74.279 pm
H-N-H Bond Angle: 180°
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 0.7428 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
H2 |
The optimisation file is linked to here
Vibrations
No negative frequencies
Molecular orbitals
H-H occupied sigma bonding orbital. 1s orbitals of H are the AOs that contribute to the MO. It is also the highest occupied molecular orbital.
H-H unoccupied sigma anti bonding orbital. Contributing AOs are the 1s orbitals of H. It is the lowest unoccupied molecular orbital.
Determination of energy of the reaction N2 + 3H2 => 2NH3
E(NH3)= -56.55776873 au 2*E(NH3)= -113.1155275 au E(N2)= -109.52412868 au E(H2)= -1.17853936 au 3*E(H2)= -3.53561808 au ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.05578074 au
ΔE=-146.452344026 kJ/mol (1au=2625.5002kJ/mol)
Since ΔE is negative the product is more stable than the gaseous reactants.
CH3OH
Key Information
Molecule: CH3OH
Calculation Method: RB3LYP
Basis Set: 6-31G(d.p)
E(RB3LYP): -115.72396437 a.u.
RMS Gradient: 0.00001481 a.u.
Point Group: C1
C-H Bond Length: 110.055 pm
C-O Bond Length: 141.811 pm
H-C-H Bond Angle: 107.89945°
C-O-H Bond Angle: 107.73743°
H-C-O Bond Angle: 112.82777°
Item Value Threshold Converged?
Maximum Force 0.000038 0.000450 YES
RMS Force 0.000020 0.000300 YES
Maximum Displacement 0.000307 0.001800 YES
RMS Displacement 0.000147 0.001200 YES
Predicted change in Energy=-1.416135D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.093 -DE/DX = 0.0 !
! R2 R(1,3) 1.1006 -DE/DX = 0.0 !
! R3 R(1,4) 1.1006 -DE/DX = 0.0 !
! R4 R(1,5) 1.4181 -DE/DX = 0.0 !
! R5 R(5,6) 0.9652 -DE/DX = 0.0 !
! A1 A(2,1,3) 107.8995 -DE/DX = 0.0 !
! A2 A(2,1,4) 107.8995 -DE/DX = 0.0 !
! A3 A(2,1,5) 106.9041 -DE/DX = 0.0 !
! A4 A(3,1,4) 108.2583 -DE/DX = 0.0 !
! A5 A(3,1,5) 112.8278 -DE/DX = 0.0 !
! A6 A(4,1,5) 112.8278 -DE/DX = 0.0 !
! A7 A(1,5,6) 107.7374 -DE/DX = 0.0 !
! D1 D(2,1,5,6) 179.9999 -DE/DX = 0.0 !
! D2 D(3,1,5,6) 61.5463 -DE/DX = 0.0 !
! D3 D(4,1,5,6) -61.5465 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
CH3OH |
The optimisation file is linked to here
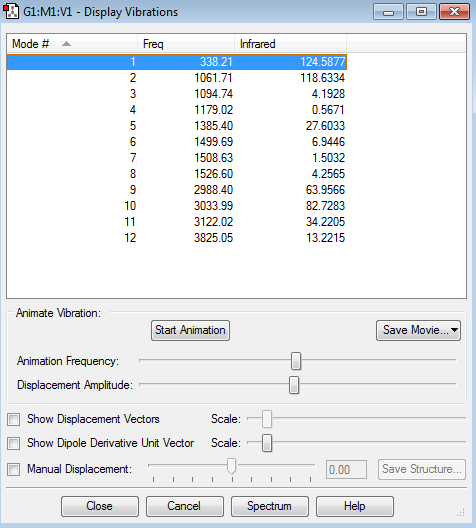
Vibrations
Expected number of modes: 12
Degenerate modesː None
Bending Vibrationsː 1,2,3,4,5,6,7,8
Bond Stretchesː 9,10,11,12
The Umbrella on is 6
Expected number of bands in a spectrum is 12
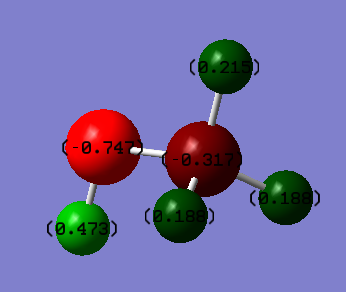
Charges
Charge on O = -0.747
Charge on H of OH = 0.473
Charge on C = -0.317
Charge on H on CH3 = 0.188
Charge on H on CH3 = 0.188
Charge on H on CH3 = 0.215
Molecular Orbitals
C-O, (occupied) sigma bonding orbital. These MOs are deep in energy. They make up the sigma bond between the atoms. The contributing AOs are the 2s orbitals of C and O. Due to the higher contribution of 2s from O, the MO is more rounded near O.
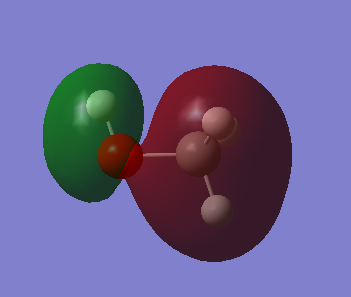
C-O, (occupied) sigma* anti bonding orbital. Contributing AOs are from 2s of C and O and the C AO contribute higher seen in picture. These MOs are deep in energy.
C-O (occupied) pi bonding orbital. 2p orbitals of the C and O are the contributing AOs. These MOs are deep in energy.
C-O pi* anti-bonding orbital which is also the highest occupied molecular orbital. The AOs are the p orbitals of C and O.
Lowest unoccupied molecular orbital. It is very hard to see what the contributing AOs are.