Rep:Mod:cwjhunt
Third year inorganic computational chemistry
Cameron Jellett
Using the computational chemistry software packages Gaussian and Gaussview, main group molecules will be created, their structures optimised, stretching frequencies calculated and their molecular orbitals visualised and analysed. Using knowledge of main group chemistry the results will be interpreted and the merit and usefulness of computational methods will be conveyed.
Optimisation of BH3
Optimisation of bond lengths and angles
The OPT optimisation for a BH3 molecule was carried out using an initial B-H bond length of 1.18000A and 120 degrees, using the B3LYP method and a 3-21G basis set, giving and optimised bond angle of 120 degrees and an optimised bond length 1.19453A of the following results: File:CWJBH3 321G.LOG

| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 3-21G |
| Charge | 0 |
| Spin | Singlet |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 3-21G |
| Charge | 0 |
| Spin | Singlet |
| E(RB3LYP) | -26.46226433a.u. |
| RMS Gradient Norm | 0.00004507a.u. |
| Dipole Moment | 0.0000Debye |
| Point Group | D3H |
| Job cpu time | 10.0 seconds. |
Item Value Threshold Converged?
Maximum Force 0.000090 0.000450 YES
RMS Force 0.000059 0.000300 YES
Maximum Displacement 0.000352 0.001800 YES
RMS Displacement 0.000230 0.001200 YES
Predicted change in Energy=-4.580970D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1945 -DE/DX = -0.0001 !
! R2 R(1,3) 1.1945 -DE/DX = -0.0001 !
! R3 R(1,4) 1.1945 -DE/DX = -0.0001 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Using the 6-31G(d,p) basis set, an optimisation on the previous optimised structure was carried out File:CWJBH6 31G.LOG. Optimised bond length = 1.19453, bond angle=120.00
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 3-21G |
| Charge | 0 |
| Spin | Singlet |
| E(RB3LYP) | -26.61531709a.u. |
| RMS Gradient Norm | 0.00052443a.u. |
| Dipole Moment | 0.0000Debye |
| Point Group | D3H |
| Job cpu time | 13.0 seconds. |
Item Value Threshold Converged?
Maximum Force 0.000009 0.000450 YES
RMS Force 0.000006 0.000300 YES
Maximum Displacement 0.000038 0.001800 YES
RMS Displacement 0.000025 0.001200 YES
Predicted change in Energy=-5.342736D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1923 -DE/DX = 0.0 !
! R2 R(1,3) 1.1923 -DE/DX = 0.0 !
! R3 R(1,4) 1.1923 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Structural optimisation of thallium tribromide
TlBr3 with a restricted symmetry of D3h and a "very tight" tolerance of 0.0001 and and initial Tl-Br bond length of A was optimised using, to give an equilibrium bond length of 2.69000 and a predetermined bond angle of 120.000 degrees, giving an optimised bond length of 2.65095A. The literature value for this bond length is 2.5122A[1] Pseudo potentials were used due to the atoms in the molecule being of a high atomic number, allowing the non-valence electrons to be modeled. DOI:10042/23352



| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Charge | 0 |
| Spin | Singlet |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | LANL2DZ |
| Charge | 0 |
| Spin | Singlet |
| E(RB3LYP) | -91.21812851a.u |
| RMS Gradient Norm | 0.00000090a.u |
| Dipole Moment | 0.0000Debye |
| Point Group | D3H |
| Job cpu time | 57.9 seconds. |
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000014 0.001200 YES
Predicted change in Energy=-6.084107D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.651 -DE/DX = 0.0 !
! R2 R(1,3) 2.651 -DE/DX = 0.0 !
! R3 R(1,4) 2.651 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Bond and angle optimsiation of BBr3
Using the GEN basis set, the LANL2DZ basis set could be used on the heavy bromine atoms and the 6-31G basis set on the boron atom, allowing the accurate analysis of BBr3 using the appropriate basis set for each atom. Inital B-Br bond length = 2.0200A and bond angle of 120.000o

| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Charge | 0 |
| Spin | Singlet |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | Gen |
| Charge | 0 |
| Spin | Singlet |
| E(RB3LYP) | -64.43645296a.u |
| RMS Gradient Norm | 0.00000382a.u |
| Dipole Moment | 0.0000Debye |
| Point Group | D3H |
| Job cpu time | 36.3 seconds. |
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000036 0.001800 YES
RMS Displacement 0.000023 0.001200 YES
Predicted change in Energy=-4.027422D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.934 -DE/DX = 0.0 !
! R2 R(1,3) 1.934 -DE/DX = 0.0 !
! R3 R(1,4) 1.934 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Summary of results
All calculations carried out using the 6-31G(d,p) basis set or LanL2DZ basis set where appropriate.
| BH3 | BBr3 | Tl3 | |
| bond legnth(A) | 1.19231 | 1.93396 | 2.65095 |
| Bond angle | 120.00 | 120.00 | 120.00 |
Chemical bonds, and covalent bonds specifically, can be considered to be an area between two atoms where there is a high electron probability density, which is considered to be an attractive interaction between the atoms. This can be also be thought of or rationalised more visually as a constructive interference of the electron clouds (utilising the 'wave-like' properties of electrons).
The relative bond lengths are as expected for these compounds, with bonds between larger atoms being longer. This is due to several factors, one being that orbitals become more diffuse and larger as one progresses down a group, meaning the overlap is not as large and the bond is longer and not as strong. H and Br both require one electron to fill their valence shell and form sigma bonds but hydrogen uses its 1s shell to overlap with the 2p shell of the boron, whereas the bromine uses its 4p shell to overlap and form a sigma bond. The B-Br bond is also more polarised than the B-H bond due to the difference in electronegativities being larger.
Thallium and boron are both in group 13, meaning the two bromides would be expected to be similar in terms of structure, the bonds are longer for the larger central atom. This is for the same reasons as discussed for comparison of bromine and hydrogen.
The angles are all at 120 degrees to minimise repulsion. The absence of non-bonding orbitals on the central atom mean there is no deviation from the trigonal planar structure
Vibrational calculations and analysis
Vibrational analysis of BH3

Low frequencies --- -3.6018 -1.1356 -0.0054 1.3735 9.7036 9.7698
Low frequencies --- 1162.9825 1213.1733 1213.1760
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A" E' E'
Frequencies -- 1162.9825 1213.1733 1213.1760
Red. masses -- 1.2531 1.1072 1.1072
Frc consts -- 0.9986 0.9601 0.9601
IR Inten -- 92.5497 14.0545 14.0581
Atom AN X Y Z X Y Z X Y Z
1 5 0.00 0.00 0.16 0.00 0.10 0.00 -0.10 0.00 0.00
2 1 0.00 0.00 -0.57 0.00 0.08 0.00 0.81 0.00 0.00
3 1 0.00 0.00 -0.57 -0.39 -0.59 0.00 0.14 0.39 0.00
4 1 0.00 0.00 -0.57 0.39 -0.59 0.00 0.14 -0.39 0.00
VIbrational analysis of TlBr3

TlBr3 frequency File Name TlBr3_frequency File Type .log Calculation Type FREQ Calculation Method RB3LYP Basis Set LANL2DZ Charge 0 Spin Singlet E(RB3LYP) -91.21812851 a.u. RMS Gradient Norm 0.00000088 a.u. Imaginary Freq 0 Dipole Moment 0.0000 Debye Point Group D3H Job cpu time: 0 days 0 hours 0 minutes 9.0 seconds.
Low frequencies --- -3.4213 -0.0026 -0.0004 0.0015 3.9367 3.9367
Low frequencies --- 46.4289 46.4292 52.1449
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
E' E' A2"
Frequencies -- 46.4289 46.4292 52.1449
Red. masses -- 88.4613 88.4613 117.7209
Frc consts -- 0.1124 0.1124 0.1886
IR Inten -- 3.6867 3.6867 5.8466
Atom AN X Y Z X Y Z X Y Z
1 81 0.00 0.28 0.00 -0.28 0.00 0.00 0.00 0.00 0.55
2 35 0.00 0.26 0.00 0.74 0.00 0.00 0.00 0.00 -0.48
3 35 0.43 -0.49 0.00 -0.01 -0.43 0.00 0.00 0.00 -0.48
4 35 -0.43 -0.49 0.00 -0.01 0.43 0.00 0.00 0.00 -0.48
Orbital analysis of BH3
The energy analysis of BH3 was carried out using the 6-31G(d,p) basis set. A molecular orbital diagram using the LCAO method was constructed to compare to the calculated MOs, revealing an elegant correlation between the two methods and highlighting the usefulness of a qualitative MO diagram. The lobes do appear to not be as narrow for the LUMO but this is not a large difference and does not discredit the qualitative method.

BH3 energy File Name BH3_energy File Type .fch Calculation Type SP Calculation Method RB3LYP Basis Set 6-31G(D,P) Charge 0 Spin Singlet Total Energy -26.61532363 a.u. RMS Gradient Norm 0.00000000 a.u. Imaginary Freq Dipole Moment 0.0000 Debye Point Group
Ammonia
NH3 optimisation
A structure optimisation using the 6-31G(d,p) basis set was carried out on a trigonal pyramidal NH3 molecule. The initial N-H bond lengths were 1.00000A and the initial HNH bond angle was 109.471 degrees, giving optimised parameters of 1.01797A and 105.741 degrees respectively. The reference values for the bond length and angles are 1.017A and 107.8o [2]. This is an agreement to 2dp and is quite close considering the simple basis set
Optimisation log file File:NH3 OPTIMISATION cwj2.LOG

NH3 optimisation File Name NH3_optimisation File Type .log Calculation Type FOPT Calculation Method RB3LYP Basis Set 6-31G(d,p) Charge 0 Spin Singlet E(RB3LYP) -56.55776856 a.u. RMS Gradient Norm 0.00000885 a.u. Imaginary Freq Dipole Moment 1.8464 Debye Point Group C1 Job cpu time: 0 days 0 hours 0 minutes 12.0 seconds.
Item Value Threshold Converged?
Maximum Force 0.000024 0.000450 YES
RMS Force 0.000012 0.000300 YES
Maximum Displacement 0.000079 0.001800 YES
RMS Displacement 0.000053 0.001200 YES
Predicted change in Energy=-1.629729D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7413 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7486 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7479 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8631 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Frequency analysis
The vibrational analysis was carried out with the 'nosymm' keyword.
Log file File:NH3 FREQUENCY cwj.LOG

| # | animation | frequency | relative intensity |
| 1 |  |
1089.56 | 145.4398 |
| 2 |  |
1694.12 | 13.5558 |
| 3 |  |
1694.19 | 13.5559 |
| 4 |  |
3460.98 | 1.0593 |
| 5 |  |
3689.40 | 0.2700 |
| 6 |  |
3589.52 | 0.2709 |
Low frequencies --- -30.7038 -0.0017 -0.0008 0.0009 20.2701 28.2996
Low frequencies --- 1089.5562 1694.1244 1694.1865
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A A
Frequencies -- 1089.5562 1694.1244 1694.1865
Red. masses -- 1.1800 1.0644 1.0644
Frc consts -- 0.8253 1.8000 1.8001
IR Inten -- 145.4398 13.5558 13.5559
Atom AN X Y Z X Y Z X Y Z
1 7 0.12 0.00 0.00 0.00 -0.02 -0.06 0.00 0.06 -0.02
2 1 -0.53 -0.21 0.00 -0.07 -0.04 0.73 0.25 0.14 0.20
3 1 -0.53 0.11 0.18 0.25 -0.24 -0.03 -0.07 -0.62 0.40
4 1 -0.53 0.11 -0.18 -0.18 0.52 0.18 -0.18 -0.41 -0.36
Energy, population and NBO analysis of NH3
The NBO analysis has given the expected information, with a higher electron density on the nitrogen due to its high electronegativity compared to the hydrogen atoms. Information about the hybridisation and the occupancy of the bonds and MOs are also as expected, with (almost) two electrons in each N-H bond and the lone pair.

NH3 energy File Name log_72698 File Type .log Calculation Type SP Calculation Method RB3LYP Basis Set 6-31G(d,p) Charge 0 Spin Singlet E(RB3LYP) -56.55776856 a.u. RMS Gradient Norm a.u. Imaginary Freq Dipole Moment 1.8464 Debye Point Group C1 Job cpu time: 0 days 0 hours 0 minutes 11.0 seconds.
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
N 1 -1.12515 1.99982 6.11104 0.01429 8.12515
H 2 0.37505 0.00000 0.62250 0.00246 0.62495
H 3 0.37505 0.00000 0.62250 0.00246 0.62495
H 4 0.37505 0.00000 0.62249 0.00246 0.62495
=======================================================================
* Total * 0.00000 1.99982 7.97852 0.02166 10.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.99909) BD ( 1) N 1 - H 2
( 68.83%) 0.8297* N 1 s( 24.87%)p 3.02( 75.05%)d 0.00( 0.09%)
-0.0001 -0.4986 -0.0059 0.0000 -0.2910
0.0052 0.8155 0.0277 0.0000 0.0000
0.0281 0.0000 0.0000 0.0032 0.0082
( 31.17%) 0.5583* H 2 s( 99.91%)p 0.00( 0.09%)
-0.9996 0.0000 0.0072 -0.0289 0.0000
2. (1.99909) BD ( 1) N 1 - H 3
( 68.83%) 0.8297* N 1 s( 24.86%)p 3.02( 75.05%)d 0.00( 0.09%)
0.0001 0.4986 0.0059 0.0000 0.2910
-0.0052 0.4077 0.0138 0.7062 0.0240
0.0140 0.0243 0.0076 0.0033 0.0031
( 31.17%) 0.5583* H 3 s( 99.91%)p 0.00( 0.09%)
0.9996 0.0000 -0.0072 -0.0145 -0.0250
3. (1.99909) BD ( 1) N 1 - H 4
( 68.83%) 0.8297* N 1 s( 24.87%)p 3.02( 75.05%)d 0.00( 0.09%)
0.0001 0.4986 0.0059 0.0000 0.2909
-0.0052 0.4077 0.0138 -0.7062 -0.0239
0.0140 -0.0243 -0.0076 0.0033 0.0031
( 31.17%) 0.5583* H 4 s( 99.91%)p 0.00( 0.09%)
0.9996 0.0000 -0.0072 -0.0145 0.0250
4. (1.99982) CR ( 1) N 1 s(100.00%)
1.0000 -0.0002 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
5. (1.99721) LP ( 1) N 1 s( 25.38%)p 2.94( 74.52%)d 0.00( 0.10%)
0.0001 0.5036 -0.0120 0.0000 -0.8618
0.0505 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (H3N)
1. BD ( 1) N 1 - H 2 1.99909 -0.60417
2. BD ( 1) N 1 - H 3 1.99909 -0.60417
3. BD ( 1) N 1 - H 4 1.99909 -0.60416
4. CR ( 1) N 1 1.99982 -14.16768
5. LP ( 1) N 1 1.99721 -0.31756
The dissociation energy of the borane-ammonia adduct
Optimisation of structure
The ammonia-borane structure was optimised using the 6-31G(d,p) basis set and RB3LYP method to optimise the structure. Initial bond lengths for BH and NH were set to that of the uncomplexed molecules. Frequency and population calculations were also carried out, all in an identical fashion as for borane and ammonia, allowing valid comparisions to be made between their ground state energies and some figures to be calculated. Using microwave spectroscopy data for the gas phase, valid comparisons can be made[3]. (very accurate neutron diffraction data for this compound is available but the calculations carried out are assumed to be in the gas phase and for a single molecule so comparing parameters is not valid. Interestingly, the B-N bond length in the solid phase is quite a bit shorter, 1.5646A, due to intermolecular forces, which were not considered for this calculation.)[4]).
File:NH3BH3 OPTIMISATION CWJ.LOG
| atoms | initial | optimised | Microwave spectroscopy data |
| NB | 1.50025A | 1.66769A | 1.67225A |
| NH | 1.11717A | 1.01846A | 1.014A |
| BH | 1.11715A | 1.20978A | 1.216A |
| HBH | 108.192o | 113.872o | 113.8o |
| HNH | 108.196o | 107.876o | 108.7o |
NH3BH3 optimisation File Name NH3BH3_optimisation_frequency File Type .log Calculation Type FOPT Calculation Method RB3LYP Basis Set 6-31G(d,p) Charge 0 Spin Singlet E(RB3LYP) -83.22468936 a.u. RMS Gradient Norm 0.00000424 a.u. Imaginary Freq Dipole Moment 5.5647 Debye Point Group C1 Job cpu time: 0 days 0 hours 1 minutes 32.0 seconds.
Item Value Threshold Converged?
Maximum Force 0.000011 0.000015 YES
RMS Force 0.000003 0.000010 YES
Maximum Displacement 0.000023 0.000060 YES
RMS Displacement 0.000008 0.000040 YES
Predicted change in Energy=-3.637174D-10
Optimization completed.
-- Stationary point found.
frequency analysis


NH3BH3 optimisation File Name NH3BH3_FREQUENCY File Type .log Calculation Type FREQ Calculation Method RB3LYP Basis Set 6-31G(d,p) Charge 0 Spin Singlet E(RB3LYP) -83.22468919 a.u. RMS Gradient Norm 0.00000402 a.u. Imaginary Freq 0 Dipole Moment 5.5647 Debye Point Group C1 Job cpu time: 0 days 0 hours 0 minutes 39.0 seconds.
Low frequencies --- -22.4465 -0.0013 -0.0010 -0.0008 5.2919 15.8327
Low frequencies --- 262.7181 633.0640 638.0599
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A A
Frequencies -- 262.7166 633.0637 638.0599
Red. masses -- 1.0078 4.9929 1.0452
Frc consts -- 0.0410 1.1790 0.2507
IR Inten -- 0.0000 13.9852 3.5550
Atom AN X Y Z X Y Z X Y Z
1 1 0.00 -0.36 -0.07 0.31 0.00 0.03 -0.17 0.13 0.07
2 1 0.00 0.24 -0.28 0.28 -0.02 -0.03 0.45 0.10 0.05
3 1 0.00 0.12 0.34 0.28 0.03 -0.02 -0.28 0.10 0.09
4 1 0.00 -0.44 -0.08 -0.34 0.00 0.00 -0.22 0.18 0.10
5 1 0.00 0.15 0.42 -0.37 0.00 -0.01 -0.36 0.16 0.12
6 1 0.00 0.29 -0.34 -0.37 0.00 -0.01 0.58 0.15 0.09
7 5 0.00 0.00 0.00 0.48 0.00 0.00 0.00 -0.02 -0.02
8 7 0.00 0.00 0.00 -0.36 0.00 0.00 0.00 -0.04 -0.03
Energy analysis
NH3BH3 optimisation File Name NH3BH3_ENERGY File Type .log Calculation Type SP Calculation Method RB3LYP Basis Set 6-31G(d,p) Charge 0 Spin Singlet E(RB3LYP) -83.22468919 a.u. RMS Gradient Norm a.u. Imaginary Freq Dipole Moment 5.5647 Debye Point Group C1 Job cpu time: 0 days 0 hours 0 minutes 7.0 seconds.
| Molecule | energy (a.u) |
| NH3 | -56.55776856 |
| BH3 | -26.61531709 |
| NH3BH3 | -83.22468919 |
Using the energy values obtained for BH3, NH3 and NH3BH3, the association/dissociation energy can be found. It is important that the values from identical basis sets and methods are used, in this case the 6-31G(d,p) basis set and RB3LYP method.
The association energy is equal to the ground state energy of the adduct minus the sum of the ground state energy of the borane and ammonia.
Association energy= -83.22468919-(-26.61531709-56.55776856)a.u. =-0.05160354a.u. Conversion of a.u. to Kjmol-1 = a.u.*2625.50=-0.0516035*2625.50 =-135.48509427 Kjmol-1
Since borane does not exist in its monomeric form without being coordinated, it is difficult to compare the calculated energy with experimental reference values, as they do not proceed via a direct reaction in the gas phase
Ionic liquid cations: onium ions
Ionic liquids are ionic compounds that have a very low melting point, rendering them liquids at room temperature. Due to the almost infinite number of combinations available to synthesise them, computational methods are used in order to try and make predictions about the properties of a given ionic liquid.
Imidazolium derivatives are the cation of choice for the best performing ionic liquids but for the purpose of this experiment simple ions will be considered. Simple alkyl Ammonium, sulphonium and phosphonium ions are all possible choices for the synthesis and their structures and MOs will be analysed and interpreted
Tetramethylammonium
Optimisation


| atoms | initial | optimised |
| CN | 1.51404A | 1.50942A |
| CH | 1.11646A | 1.09017A |
| CNC | 108.306o | 109.471o |
File Name ammonium_optimisation File Type .log Calculation Type FOPT Calculation Method RB3LYP Basis Set 6-31G(d,p) Charge 1 Spin Singlet E(RB3LYP) -214.18127245 a.u. RMS Gradient Norm 0.00000318 a.u. Imaginary Freq Dipole Moment 0.0000 Debye Point Group C1 Job cpu time: 0 days 0 hours 32 minutes 58.9 seconds.
Item Value Threshold Converged?
Maximum Force 0.000007 0.000015 YES
RMS Force 0.000002 0.000010 YES
Maximum Displacement 0.000049 0.000060 YES
RMS Displacement 0.000017 0.000040 YES
Predicted change in Energy=-7.577346D-12
Optimization completed.
-- Stationary point found.
Frequency analysis
File Name ammonium_optimisation File Type .log Calculation Type FOPT Calculation Method RB3LYP Basis Set 6-31G(d,p) Charge 1 Spin Singlet E(RB3LYP) -214.17522293 a.u. RMS Gradient Norm 0.00943397 a.u. Imaginary Freq Dipole Moment 0.0091 Debye Point Group C1 Job cpu time: 0 days 0 hours 32 minutes 58.9 seconds.
Low frequencies --- -8.8708 -0.0004 0.0007 0.0009 3.1678 7.3859
Low frequencies --- 182.6329 288.5463 288.8383
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A A
Frequencies -- 182.6328 288.5462 288.8383
Red. masses -- 1.0078 1.0331 1.0331
Frc consts -- 0.0198 0.0507 0.0508
IR Inten -- 0.0000 0.0000 0.0000
=== Energy and NBO analysis ===
Ammonium energy File Name ammonium_energy File Type .fch Calculation Type SP Calculation Method RB3LYP Basis Set 6-31G(D,P) Charge 1 Spin Singlet Total Energy -214.18127340 a.u. RMS Gradient Norm 0.00000000 a.u. Imaginary Freq Dipole Moment 0.0000 Debye Point Group

| atom | charge |
| C | -0.483 |
| N | -0.295 |
| H | 0.269 |
Tetramethyl phosphonium
Optimisation


| atoms | initial | optimised |
| CP | 1.51404A | 1.81638A |
| CH | 1.11646A | 1.09329A |
| CPC | 110.603o | 109.472o |
tetramethylphosphonium optimisation File Name phosphonium optimisation File Type .log Calculation Type FOPT Calculation Method RB3LYP Basis Set 6-31G(d,p) Charge 1 Spin Singlet E(RB3LYP) -500.82701012 a.u. RMS Gradient Norm 0.00000459 a.u. Imaginary Freq Dipole Moment 0.0000 Debye Point Group C1 Job cpu time: 0 days 0 hours 54 minutes 17.6 seconds.
Item Value Threshold Converged?
Maximum Force 0.000013 0.000015 YES
RMS Force 0.000003 0.000010 YES
Maximum Displacement 0.000050 0.000060 YES
RMS Displacement 0.000016 0.000040 YES
Predicted change in Energy=-1.199087D-10
Optimization completed.
-- Stationary point found.
Frequency analysis
tetramethylphosphonium frequency File Name phosphonium frequency File Type .log Calculation Type FREQ Calculation Method RB3LYP Basis Set 6-31G(d,p) Charge 1 Spin Singlet E(RB3LYP) -500.82701069 a.u. RMS Gradient Norm 0.00000528 a.u. Imaginary Freq 0 Dipole Moment 0.0000 Debye Point Group C1 Job cpu time: 0 days 0 hours 20 minutes 37.2 seconds.
Low frequencies --- -9.8943 -6.0983 -0.0025 -0.0024 -0.0020 9.8062
Low frequencies --- 155.9463 190.9155 191.6684
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A A
Frequencies -- 155.9456 190.9135 191.6683
Red. masses -- 1.0078 1.0255 1.0255
Frc consts -- 0.0144 0.0220 0.0222
IR Inten -- 0.0000 0.0000 0.0000
Energy and NBO analysis
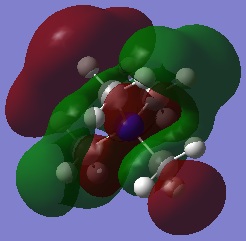
| Visualisation | Energy(a.u) | |
| HOMO | 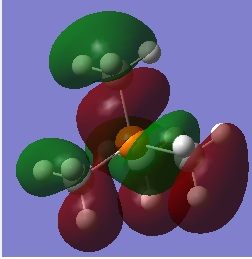 |
-0.53298 |
| LUMO | 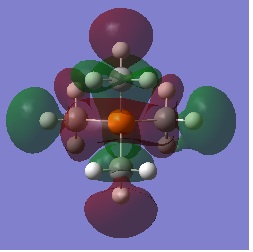 |
-0.11004 |
| NBO | 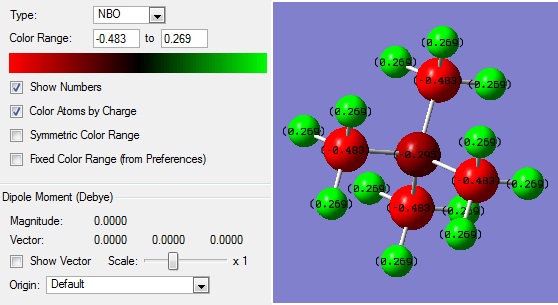 |
- |
tetramethylphosphonium energy File Name phosphonium_energy File Type .fch Calculation Type FREQ Calculation Method RB3LYP Basis Set 6-31G(D,P) Charge 1 Spin Singlet Total Energy -500.82701069 a.u. RMS Gradient Norm 0.00000528 a.u. Imaginary Freq Dipole Moment 0.0000 Debye Point Group
Trimethylsulphonium
Optimisation
The structure was drawn and the "clean" button was used to give a rough optimised structure, changing the structure from an initial trigonal planar structure to a trigonal pyramidal structure.


| atoms | initial | optimised |
| CS | 1.78000A | 1.82263A |
| CH | 1.07000A | 1.09195A |
| CSC | 109.126o | 102.748o |
sulphonium optimisation File Name sulphonium_optimisation File Type .log Calculation Type FOPT Calculation Method RB3LYP Basis Set 6-31G(d,p) Charge 1 Spin Singlet E(RB3LYP) -517.67146472 a.u. RMS Gradient Norm 0.01337505 a.u. Imaginary Freq Dipole Moment 2.4542 Debye Point Group C3V Job cpu time: 0 days 0 hours 9 minutes 45.1 seconds.
Item Value Threshold Converged?
Maximum Force 0.000000 0.000015 YES
RMS Force 0.000000 0.000010 YES
Maximum Displacement 0.000042 0.000060 YES
RMS Displacement 0.000015 0.000040 YES
Predicted change in Energy=-1.715935D-11
Optimization completed.
-- Stationary point found.
Frequency analysis
Early attempts for this calculation led to very large low frequency values (-187! DOI:10042/23597 ) with the integral=grid=ultrafine key words. When the optimisation and frequency analysis was carried out using the "nosymm" keyword, these issues were resolved
sulphonium optimisation File Name sulphonium_frequency File Type .log Calculation Type FREQ Calculation Method RB3LYP Basis Set 6-31G(d,p) Charge 1 Spin Singlet E(RB3LYP) -517.68327433 a.u. RMS Gradient Norm 0.00000059 a.u. Imaginary Freq 0 Dipole Moment 2.6179 Debye Point Group C1 Job cpu time: 0 days 0 hours 10 minutes 1.7 seconds.
Low frequencies --- -5.5886 -0.0041 -0.0038 -0.0036 2.0026 4.7146 Low frequencies --- 161.9128 199.8413 200.5485
Energy
sulphonium optimisation File Name sulphonium_energy File Type .log Calculation Type SP Calculation Method RB3LYP Basis Set 6-31G(d,p) Charge 1 Spin Singlet E(RB3LYP) -517.68327433 a.u. RMS Gradient Norm a.u. Imaginary Freq Dipole Moment 2.6179 Debye Point Group C1 Job cpu time: 0 days 0 hours 1 minutes 28.5 seconds.
| Visualisation | Energy(a.u) | |
| HOMO | 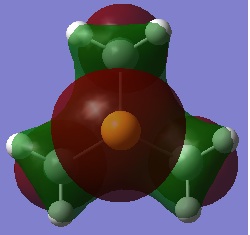 |
-0.51512 |
| LUMO | 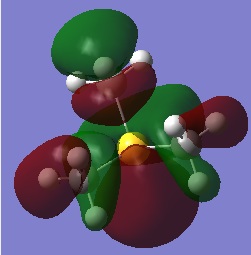 |
-0.17626 |
| NBO | 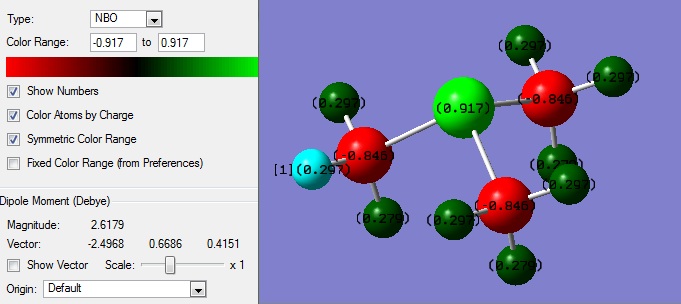 |
- |
Summary: Comparing onium ions
The liquid state of such salts is due to complex intramolecular ionic interactions. One reason for their low melting point is the formation of ion-pairs, which screen charge and reduce the attractive force between ions, localising the colombic interaction. There are also other very complicated dipole-dipole interactions and dipole-ion interactions which are beyond the scope of these calculations. The reference also mentions that asymmetry reduces these culombic attractions.[5] From the charge distribution data, some interesting differences can be found between the molecules. Comparisons down a group (nitrogen and phosphorous) and across a period (phosphorous and sulphur) can be made, their relative electronegativities and size being considered. The charges on the atoms of the ions is somewhat in agreement with their electronegativities, with the majority of the positive charge lying on the hydrogen atoms in each case. However, the central atom charge difference between the carbons is not in agreement, with a higher positive charge than expected in each case.
Trimethylhydroxylmethylammonium ion
Optimisation


Title Card Required File Name NOH_optimisation_cwj File Type .log Calculation Type FOPT Calculation Method RB3LYP Basis Set 6-31G(d,p) Charge 1 Spin Singlet E(RB3LYP) -289.39321740 a.u. RMS Gradient Norm 0.00000130 a.u. Imaginary Freq Dipole Moment 1.5785 Debye Point Group CS Job cpu time: 0 days 0 hours 16 minutes 38.3 seconds.
Item Value Threshold Converged?
Maximum Force 0.000003 0.000015 YES
RMS Force 0.000001 0.000010 YES
Maximum Displacement 0.000035 0.000060 YES
RMS Displacement 0.000010 0.000040 YES
Predicted change in Energy=-8.844343D-11
Optimization completed.
-- Stationary point found.
Frequency analysis
NOH frequency File Name NOH_frequency2_cwj File Type .log Calculation Type FREQ Calculation Method RB3LYP Basis Set 6-31G(d,p) Charge 1 Spin Singlet E(RB3LYP) -289.39470635 a.u. RMS Gradient Norm 0.00000125 a.u. Imaginary Freq 0 Dipole Moment 4.7782 Debye Point Group C1 Job cpu time: 0 days 0 hours 28 minutes 7.0 seconds.
Low frequencies --- -11.5812 -4.5952 -0.0013 -0.0010 -0.0008 3.0716
Low frequencies --- 130.7595 214.4481 255.5359
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A A
Frequencies -- 130.7569 214.4478 255.5356
Red. masses -- 2.1678 1.1196 2.7347
Frc consts -- 0.0218 0.0303 0.1052
IR Inten -- 5.1700 3.3960 27.6789
MO and energy analysis
NOH frequency File Name energy File Type .log Calculation Type SP Calculation Method RB3LYP Basis Set 6-31G(d,p) Charge 1 Spin Singlet E(RB3LYP) -289.39470717 a.u. RMS Gradient Norm a.u. Imaginary Freq Dipole Moment 2.1358 Debye Point Group C1 Job cpu time: 0 days 0 hours 2 minutes 56.1 seconds.
| Visualisation | Energy(a.u) | |
| HOMO |  |
-0.48763 |
| LUMO |  |
-0.12459 |
| NBO |  |
- |
Summary
Trimethylacetonitrileammonium ion
Optimisation


NR3CN File Name NR3CN_optimisation_cwj File Type .log Calculation Type FOPT Calculation Method RB3LYP Basis Set 6-31G(d,p) Charge 1 Spin Singlet E(RB3LYP) -306.39376190 a.u. RMS Gradient Norm 0.00000072 a.u. Imaginary Freq Dipole Moment 4.1224 Debye Point Group C1 Job cpu time: 0 days 0 hours 28 minutes 25.2 seconds.
Item Value Threshold Converged?
Maximum Force 0.000001 0.000015 YES
RMS Force 0.000000 0.000010 YES
Maximum Displacement 0.000054 0.000060 YES
RMS Displacement 0.000016 0.000040 YES
Predicted change in Energy=-1.064161D-10
Optimization completed.
-- Stationary point found.
Frequency analysis
NR3CN frequency File Name NR3CN_frequency_cwj File Type .log Calculation Type FREQ Calculation Method RB3LYP Basis Set 6-31G(d,p) Charge 1 Spin Singlet E(RB3LYP) -306.39376190 a.u. RMS Gradient Norm 0.00000074 a.u. Imaginary Freq 0 Dipole Moment 4.1224 Debye Point Group C1 Job cpu time: 0 days 0 hours 31 minutes 40.0 seconds.
Low frequencies --- -4.9511 -2.5804 0.0009 0.0009 0.0013 4.8385 Low frequencies --- 91.6979 153.9763 211.2379
Energy and NBO analysis
NR3CN frequency File Name NCN3_energy File Type .log Calculation Type SP Calculation Method RB3LYP Basis Set 6-31G(d,p) Charge 1 Spin Singlet E(RB3LYP) -306.39376190 a.u. RMS Gradient Norm a.u. Imaginary Freq Dipole Moment 4.1224 Debye Point Group C1 Job cpu time: 0 days 0 hours 3 minutes 22.6 seconds.
| Visualisation | Energy(a.u) | |
| HOMO | 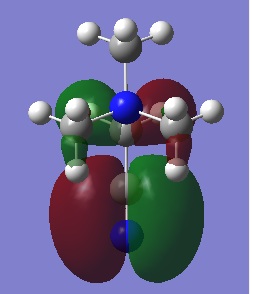 |
-0.48763 |
| LUMO |  |
-0.12459 |
| NBO | 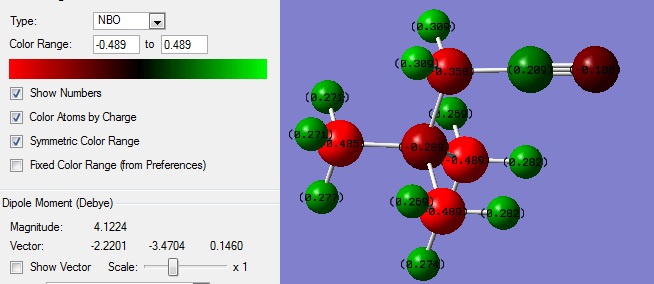 |
- |
Summary
HOMO-LUMO gap = 0.61222a.u
References
- ↑ J.Glaser, G.Johansson, Acta Chemica Scandanavica, 1982, 36A, 125-35
- ↑ Linstrom, P.J.; Mallard, W.G. (eds.) NIST Chemistry WebBook, NIST Standard Reference Database Number 69. National Institute of Standards and Technology, Gaithersburg MD
- ↑ Suenram, R. D., and F. J. Lovas. "Microwave spectrum, torsional barrier, and structure of BH3NH3." The Journal of Chemical Physics 78.1 (1983): 167-171.
- ↑ Stephens, Frances H., Vincent Pons, and R. Tom Baker. "Ammonia–borane: the hydrogen source par excellence?." Dalton Transactions 25 (2007): 2613-2626.
- ↑ Weingärtner, Hermann. "Understanding ionic liquids at the molecular level: facts, problems, and controversies." Angewandte Chemie International Edition 47.4 (2007): 654-670.