Rep:Mod:cissy
Part I: Conformational Analysis using Molecular Mechanics
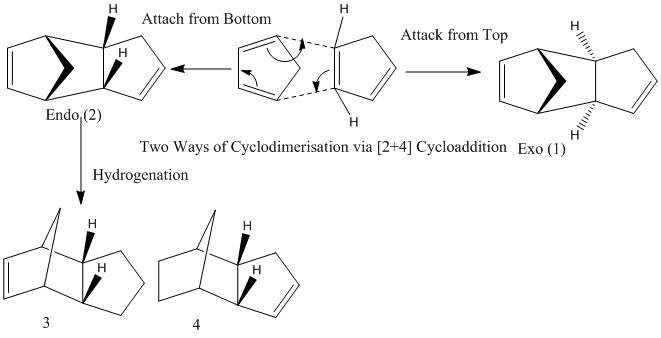
The Hydrogenation of Cyclopentadiene Dimer
| Cyclopentadiene | Exo Dimer 1 [Exo dimer file] | Endo Dimer 2 [Endo dimer file] | Dihydro derivative 3 [Dihydro 3 file] | Dihydro derivative 4 [Dihydro 4 file] |
|---|---|---|---|---|
| Total bond stretching energy kcal/mol | 3.54319 | 3.46798 | 3.31164 | 2.82311 |
| Total angle bending energy kcal/mol | 30.77256 | 33.18862 | 31.93160 | 24.68534 |
| Total Stretch Bending Energy kcal/mol | -2.04143 | -2.08221 | -2.10207 | -1.65719 |
| Total torsional energy kcal/mol | -2.73082 | -2.94983 | -1.46682 | -0.37841 |
| Total Out-of-Plane Bending Energy | 0.01476 | 0.02184 | 0.01311 | 0.00028 |
| Total van der Waals energy kcal/mol | 12.80149 | 12.35922 | 13.63874 | 10.63734 |
| Total electrostatic energy kcal/mol | 13.01367 | 14.18504 | 5.11947 | 5.14702 |
| Total Energy kcal/mol | 55.37342 | 58.19067 | 50.44569 | 41.25749 |
| Auto Optimisation Energy kJ/mol | 231.837 | 243.633 | 211.206 | 172.737 |
Comment:
cyclodimerisation of cycyclopentadiene


The calculation is implemented in Avogadro program using MMFF94s force field. Same force field is applied in the following calculations to allow comparisons. The table above shows the exo isomer has a lower energy (55.37342 kcal/mol) than endo isomer (58.19067 kcal/mol). It implies it should give an exo molecule if the dimerisation is under thermodynamic control. However, more endo dimers are observed in reality.
As can be seen from the structure, there is less steric hindrance in exo, because the an eclipsed conformation to the cyclopentadiene ring. However as the two C=C double bond can communicate more easily in Endo structure, this feature stabilies structure and favours kinetically controlled endo isomer. The bigger value in total angle bending energy, total stretch bending energy, total torsional energy and total electrostatic energy indicates a larger deviation from 'normality', especially in angle bending energy. The ideal hybridisation angles are sp2 C: 120°, sp3 C: 109.5°.
As in reality, it forms endo dimer 2 rather than dimer 1. Therefore the cyclodimerisation of cycyclopentadiene is kinetically controlled due to the relatively small barrier. The diagram of the frontier approach is shown on the right.
Hydrogenation of dimer
Dihydro derivative 3/4 are obtained by hydrogenating the left and right C=C double bond of endo dimer 2 respectively. dihydro derivative 4 is more stable and about 9 kcal/mol less in total tthan derivative 3. The contribution of each component is that there are less Stretching, bending and van der waals energy in derivative 4. But the torsion is stabilises more in derivative 3 than derivative 4. Although electrostatic energy is slightly higher in derivative 4, overall, derivative 4 is thermodynamically more favoured.
The Intermediate of Taxol
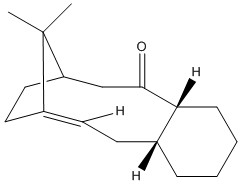
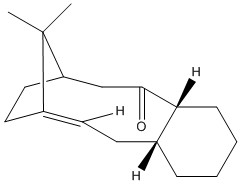
| Intermediate | Intermediate 9 (Chair 1) [Jmol file of 9 chair 1] | Intermediate 9 (Chair 2) [Jmol file of 9 chair 2] | Intermediate 10 (Chair 1) [Jmol file of 10 (Chair 1)] | Intermediate 10 (Chair 2) [Jmol file of 10 (Chair 2)] | Intermediate 10 (Twisted boat) [Jmol file of 10 (twisted boat)] |
|---|---|---|---|---|---|
| Total bond stretching energy kcal/mol | 7.65698 | 8.55713 | 7.59784 | 8.89690 | 7.75526 |
| Total angle bending energy kcal/mol | 28.25773 | 33.49381 | 18.79658 | 22.92290 | 19.01147 |
| Total Stretch Bending Energy kcal/mol | -0.08444 | 0.07636 | -0.14229 | -0.15202 | -0.13458 |
| Total torsional energy kcal/mol | 0.28713 | 3.37588 | 0.21725 | 4.32995 | 3.76443 |
| Total Out-of-Plane Bending Energy | 0.97415 | 1.46318 | 0.84050 | 1.65787 | 0.95709 |
| Total van der Waals energy kcal/mol | 33.14864 | 35.83975 | 33.29621 | 36.86870 | 35.00206 |
| Total electrostatic energy kcal/mol | 0.30182 | 0.23331 | -0.05266 | 0.44811 | -0.06146 |
| Total Energy kcal/mol | 70.54201 | 83.03941 | 60.55342 | 74.97241 | 66.29428 |
| Auto Optimisation Energy kJ/mol | 295.345 | 347.669 | 253.525 | 313.894 | 277.561 |
Comment:
Same force field (MMFF94s) is used to allow the comparison. As can be seen, Intermediate 10 is relatively more stable in most of its conformations. The chair 1 form is the most stable conformation in both cases. The twisted boat form of 10 is also more stable than chair 2. Given the most ideal optimised conformations, a more thermodynamically stable Intermediate 10 (chair 1) is observed by approximately 10 kcal lower than intermediate 9 (chair 1).
| Hydrogenation product | product 9 [Jmol file of product 9] | Product 10 [Jmol file of product 10] |
|---|---|---|
| Total bond stretching energy kcal/mol | 6.95253 | 6.57499 |
| Total angle bending energy kcal/mol | 32.06881 | 24.75416 |
| Total Stretch Bending Energy kcal/mol | 0.30114 | 0.42789 |
| Total torsional energy kcal/mol | 9.47305 | 8.46687 |
| Total Out-of-Plane Bending Energy | 0.24829 | 0.06434 |
| Total van der Waals energy kcal/mol | 32.71131 | 31.13884 |
| Total electrostatic energy kcal/mol | 0.00000 | 0.00000 |
| Total Energy kcal/mol | 81.75514 | 71.42709 |
| Auto Optimisation Energy kJ/mol | 342.292 | 299.051 |
Comment:
Why the alkene reacts slowly?[1]
Olefin Strain Energy is the total torsional energy difference between an olefin and its hydrogated derivative. Therefore it is 9.18592 kcal/mol (9.47305-0.28713 kcal/mol) for bridgehead olefin isomer 9 and 8.24962 kcal/mol (8.46687- 0.21725 kcal/mol) for isomer 10. As their Olefin strain energy are both lower than 17 kcal/mol, they are both kinetically stable at room temperature. It consequences the slow reaction of these alkenes.
Spectroscopic Simulation using Quantum Mechanics
| File Name | derivative 18b | derivative 18boat | |
|---|---|---|---|
| File Type | .fch | .fch | |
| Calculation Type | FREQ | FREQ | |
| Calculation Method | RB3LYP | RB3LYP | |
| Basis Set | 6-31G(D,P) | 6-31G(D,P) | |
| Change | 0 | 0 | |
| Spin | Singlet | Singlet | |
| E(RB3LYP) | -1651.88339390 | -1651.87982486 | a.u. |
| RMS Gradient Norm | 0.00000675 | 0.00001639 | a.u. |
| Imaginary Freq | |||
| Dipole Moment | 1.8267 | 1.6490 | Debye |
| Point Group |
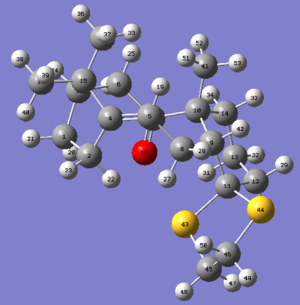
The NMR calculation is run for derivative 18 in both chair and twisted boat conformation, using MMFF94s mechanics force field, B3LYP/ 6-31G(d,p). The geometry is minimised and optimised, by Avogadro, calculated by HPC and output by Gaussian. The solvent used is benzene so as to compare the literature value (C6D6) more accurately. The chemical shifts of the hydrogens on three methyl groups are averaged before comparison. The data are summarised and analysed in the tables below and the original summaries are attached following as well.
1H NMR Table
H NMR Chair Conformation Boat Conformation Litt. Average Multi. Integral δ(ppm) Dev. δ(ppm) Dev. Atoms 5.21 5.21 m 1.00 5.97 0.76 5.46 0.25 19 3.00-2.70 2.85 m 6.00 2.70 -0.15 2.47 -0.37 27, 28, 25, 22, 20, 31 2.70-2.35 2.525 m 4.00 3.02 0.50 3.03 0.51 47, 48, 49, 50 2.20-1.70 1.95 m 6.00 1.91 -0.04 1.86 -0.09 30, 23, 24, 29, 21, 26 1.58 1.58 t 1.00 2.53 0.95 2.63 1.05 42 1.50-1.20 1.35 m 3.00 1.49 0.14 1.45 0.10 32, 33, 34 1.1 1.1 s 3.00 1.40 0.30 1.43 0.33 51, 52, 53 1.07 1.07 s 3.00 1.27 0.20 1.26 0.19 35, 36, 37 1.03 1.03 s 3.00 1.13 0.10 1.16 0.13 38, 39, 40 Sum of difference for Chair Conformation: 3.08 Average Absolute Deviation for Chair Conformation: 0.308 Sum of difference for Boat Conformation: 2.32 Average Absolute Deviation for Boat Conformation: 0.232
In chair conformation of derivative 18, H19, H42, and Hydrogen on three methyl groups are accurately assigned. The higher chemical shift of methyl group (H51,52,53) than literature may be due to loss of correction of the Nuclear Overhauser Effect(NOE) between the syn-oriented bridgehead methyl to carbonyl. The rest is matched and averaged in orders. As can be seen from the table, the chemical shifts from calculation are generally higher than the ones in literature apart from the two 6H multiplicities. In twisted boat conformation, it matches better than chair form as it has a smaller average absolute value.
13C NMR Table
Chair Conformation Boat Conformation
Lit. Calc. Diff. Atom Shift (ppm) Atom
211.49 211.93 0.44 7 209.0818827 -2.41 7
148.72 147.87 -0.85 4 148.8490003 0.13 4
120.90 120.12 -0.78 5 118.593708 -2.31 5
74.61 92.84 18.23 11 91.12128445 16.51 11
60.53 65.94 5.41 9 64.46792615 3.94 9
51.30 54.94 3.64 3 55.17891048 3.88 3
50.94 54.76 3.82 10 54.56777452 3.63 10
45.53 49.53 4.00 15 49.95651558 4.43 15
43.28 48.03 4.75 12 45.67645398 2.40 46
40.82 45.65 4.83 45 41.77107157 0.95 45
38.73 44.01 5.28 46 37.82825062 -0.90 12
36.78 41.47 4.69 14 37.55686095 0.78 6
35.47 38.51 3.04 6 35.54228625 0.07 14
30.84 33.69 2.85 41 34.10497949 3.26 8
30.00 32.47 2.47 8 32.18020025 2.18 41
25.56 28.36 2.80 2 28.51957934 2.96 2
25.35 26.50 1.15 17 26.14009992 0.79 17
22.21 24.44 2.23 1 25.05070644 2.84 1
21.39 24.01 2.62 13 22.86002242 1.47 16
19.83 22.58 2.75 16 22.28061112 2.45 13
Sum of difference for Chair Conformation: 73.36
Average Absolute Deviation for Chair Conformation: 3.668
Sum of difference for Boat Conformation: 47.05
Average Absolute Deviation for Boat Conformation: 2.352
The calculated value is recorded as a descend order in terms of chemical shift (the original summaries are attached as below) so as to compare the literature value. In general, in both conformation, the chemical shift match quite well at high chemical shift. The chair conformation works better in particular at the first three chemical shift above 100 ppm. The greatest deviation between the predicted value and literature value is at C11 with more than 15 ppm difference in both cases. After that, the average deviation in chair conformation is 3.52 ppm and 2.20 ppm in twisted boat. This indicates are better predication in twisted boat than in chair conformation. The deviation may be caused by not enough corrections. Especially for C11, the spin-orbit correction error results in a large deviation from the literature value. Overall, the sum of difference in each case gives 73.36 ppm in chair and 47.05 ppm in twisted boat with average absolute deviation 3.668 ppm and 2.352 ppm respectively. Therefore, overall the twisted boat conformation works better for prediction of 13C NMR spectrum.
Another good method doing this is the average of each squared deviation to see how far it is from zero. This is different from standard deviation which to look at how far it is from the average.
Free energy ΔG The Sum of electronic and thermal Free Energies are -1651.463261, -1651.458871, -1651.460773 respectively in derivative 18 chair, twisted boat and derivative 17 chair. As can be seen, derivative 18 is thermodynamically more stable.
SVG for Derivative 18 H NMR, chair
# Summary of NMR spectra (SCF GIAO Magnetic shielding)
# Values for element H only
# Reference: TMS B3LYP/6-31G(d,p) Benzene
# Reference shielding: 31.7499 ppm
# Degenerate peaks are condensed together (Degeneracy Tolerance 0.001)
#
# Shift (ppm) Degeneracy Atoms
5.9728187228 1.0000 19
3.1468330726 1.0000 47
3.0997002105 1.0000 50
2.9569032079 1.0000 48
2.9484932393 1.0000 27
2.8941977150 1.0000 49
2.8222703834 1.0000 28
2.7750858497 1.0000 25
2.6719690758 1.0000 22
2.5556737278 1.0000 20
2.5271749699 1.0000 42
2.4263461049 1.0000 31
2.3321587488 1.0000 51
2.2972563381 1.0000 30
1.9969977089 1.0000 23
1.9640755030 1.0000 24
1.8465941231 1.0000 29
1.8085551544 1.0000 21
1.6399309967 1.0000 35
1.5749611497 1.0000 26
1.5329369739 1.0000 40
1.4959885397 1.0000 34
1.3400344766 1.0000 32
1.2733940992 1.0000 52
1.2117595975 1.0000 33
1.2046295252 1.0000 37
0.9678573732 1.0000 39
0.9590007145 1.0000 36
0.8995562458 1.0000 38
0.6010468880 1.0000 53
SVG for Derivative 18 C NMR, chair
# Summary of NMR spectra (SCF GIAO Magnetic shielding)
# Values for element C only
# Reference: TMS B3LYP/6-31G(d,p) Benzene
# Reference shielding: 192.066 ppm
# Degenerate peaks are condensed together (Degeneracy Tolerance 0.001)
#
# Shift (ppm) Degeneracy Atoms
211.9254849043 1.0000 7
147.8680072496 1.0000 4
120.1200682015 1.0000 5
92.8437310860 1.0000 11
65.9407393198 1.0000 9
54.9353476663 1.0000 3
54.7554216364 1.0000 10
49.5291537658 1.0000 15
48.0311731199 1.0000 12
45.6518513246 1.0000 45
44.0071543232 1.0000 46
41.4699312581 1.0000 14
38.5109384763 1.0000 6
33.6910774841 1.0000 41
32.4730289109 1.0000 8
28.3566420906 1.0000 2
26.5012997733 1.0000 17
24.4402611433 1.0000 1
24.0062973991 1.0000 13
22.5826199650 1.0000 16
SVG for Derivative 18 H NMR, boat
# Summary of NMR spectra (SCF GIAO Magnetic shielding)
# Values for element H only
# Reference: TMS B3LYP/6-31G(d,p) Benzene
# Reference shielding: 31.7499 ppm
# Degenerate peaks are condensed together (Degeneracy Tolerance 0.05)
#
# Shift (ppm) Degeneracy Atoms
5.4614288057 1.0000 19
3.2172500183 1.0000 50
3.0931425370 1.0000 49
2.9203783083 2.0000 47,48
2.7818553570 1.0000 25
2.6803702579 1.0000 30
2.6262739114 1.0000 42
2.4806466526 1.0000 20
2.3639930206 2.0000 28,27
2.1862862498 1.0000 22
2.0013384220 2.0000 24,23
1.8733826516 2.0000 51,21
1.7623387895 3.0000 29,31,32
1.5424814315 4.0000 35,33,40,26
1.2745207736 2.0000 34,52
1.1634079006 2.0000 37,53
1.0091051265 2.0000 39,36
0.9342856961 1.0000 38
SVG for Derivative 18 C NMR, boat
# Summary of NMR spectra (SCF GIAO Magnetic shielding)
# Values for element C only
# Reference: TMS B3LYP/6-31G(d,p) Benzene
# Reference shielding: 192.066 ppm
# Degenerate peaks are condensed together (Degeneracy Tolerance 0.05)
#
# Shift (ppm) Degeneracy Atoms
209.0818826768 1.0000 7
148.8490002954 1.0000 4
118.5937079920 1.0000 5
91.1212844452 1.0000 11
64.4679261514 1.0000 9
55.1789104843 1.0000 3
54.5677745152 1.0000 10
49.9565155846 1.0000 15
45.6764539827 1.0000 46
41.7710715700 1.0000 45
37.8282506154 1.0000 12
37.5568609474 1.0000 6
35.5422862538 1.0000 14
34.1049794907 1.0000 8
32.1802002478 1.0000 41
28.5195793411 1.0000 2
26.1400999217 1.0000 17
25.0507064423 1.0000 1
22.8600224209 1.0000 16
22.2806111235 1.0000 13
Free Energy ΔG
D-Space Derivative 18, Chair (Solvent: Chloroform)
Zero-point correction= 0.467822 (Hartree/Particle) Thermal correction to Energy= 0.489247 Thermal correction to Enthalpy= 0.490191 Thermal correction to Gibbs Free Energy= 0.421082 Sum of electronic and zero-point Energies= -1651.416521 Sum of electronic and thermal Energies= -1651.395096 Sum of electronic and thermal Enthalpies= -1651.394152 Sum of electronic and thermal Free Energies= -1651.463261
D-Space Derivative 18, Chair (Solvent: Benzene)
Zero-point correction= 0.467920 (Hartree/Particle) Thermal correction to Energy= 0.489409 Thermal correction to Enthalpy= 0.490353 Thermal correction to Gibbs Free Energy= 0.420954 Sum of electronic and zero-point Energies= -1651.411905 Sum of electronic and thermal Energies= -1651.390416 Sum of electronic and thermal Enthalpies= -1651.389472 Sum of electronic and thermal Free Energies= -1651.458871
D-Space Derivative 17, Chair (Solvent: Chloroform)
Zero-point correction= 0.468396 (Hartree/Particle) Thermal correction to Energy= 0.489751 Thermal correction to Enthalpy= 0.490695 Thermal correction to Gibbs Free Energy= 0.422008 Sum of electronic and zero-point Energies= -1651.414385 Sum of electronic and thermal Energies= -1651.393029 Sum of electronic and thermal Enthalpies= -1651.392085 Sum of electronic and thermal Free Energies= -1651.460773
Part 2: Analysis of the Properties of the synthesised Alkene Epoxides
Two Catalytic System
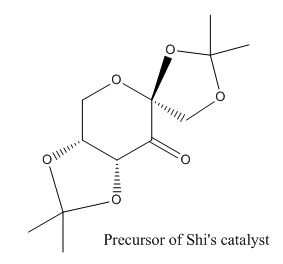
Crystal Structure of Shi's Catalyst
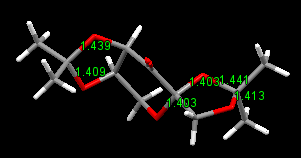
Three pairs of bond lengths of O-C-O are marked from left to right in Mercury program. The carbon of the middle pair is an anomeric centre with bond length 1.403 Å and 1.403 Å. It is the evidence of anomeric effect. In comparison, the two O-C bonds not at the anomeric centre are different in lengths and slightly longer.
The bond angle is further investigated. It turns out that the O-C-O bond angle at the anomeric centre is close to 120, sp2 like orbital. It also indicates the delocalised π system between O-C-O due to the anomeric effect, while the other two O-C-O are close to 109, tetrahedral sp3 like.
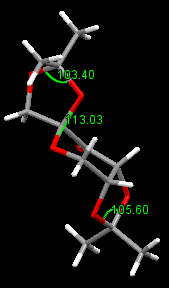
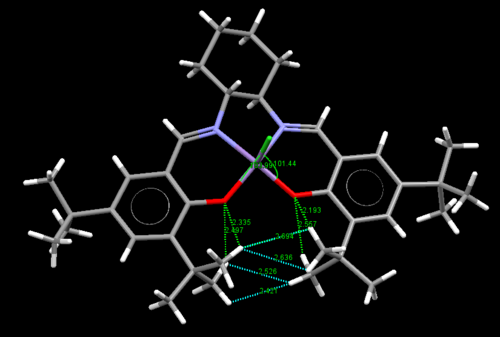
Crystal Structure of Jacobsen Catalyst
The van der Waals radii are investigated here between the two adjacent t-butyl groups. Two pairs of Hydrogens in upper layer is 2.694 and 2.636 Å. The two pairs at the bottom layer is 2.526 and 2.421 Å. The favorable Van Der Waals Interaction is between 2.1 Å and 2.4Å[3] . Therefore, H...H is quite favorable in the bottom methyl groups but slightly repulsive in the top methyl groups. Further investigation is to look at the O...H distance. it is 2.335 Å and 2.193 Å in the top methyl groups and 2.497 Å, 2.557 in the bottom methyl groups. The ideal attraction occurs when the O...H distance is lower than 2.62 Å. So O...H interaction is attractive in all four O...H pairs and stronger in top methyl groups. Therefore the O...H attraction in the top methyl groups compensate the slight repulsion between the hydrogen atoms. This overall stabilises the t-butyl groups there. In addition, the Cl-Mn-O bond angles are checked. The angles turn out to be greater than 90, which indicates the Jacobsen pre-catalyst is not flat. The slightly bent structure result in a shorter distance of H...H at the bottom methyl groups.
Epoxide NMR Analysis
Stilbene Oxide NMR Analysis
| R-Stilbene | H NMR svg | C NMR svg | S-Stilbene | H NMR svg | C NMR svg | Literature[4] | ||
|

|

|
|

|

|
H NMR (400 MHz, CDCl3) | C NMR (100 MHz, CDCl3) | |
| H-NMR | δ ppm | Degeneracy | Atom | δ ppm | Degeneracy | Atom | δ ppm | Multiplicity |
| 7.57 | 2 | 24,20 | 7.57 | 2 | 26,20 | 7.30-7.38 | m, 10H | |
| 7.48 | 8 | 18,27,17,26,25,16,19,23 | 7.48 | 8 | 18,23,17,24,25,16,19,27 | |||
| 3.54 | 2 | 21,22 | 3.54 | 2 | 21,22 | 3.87 | s,2H | |
| C NMR | 134.09 | 2 | 4,9 | 134.09 | 2 | 4,9 | 137.1 | 2C |
| 124.22 | 2 | 11,6 | 124.22 | 2 | 13,6 | 128.5 | 4CH | |
| 123.52 | 2 | 2,13 | 123.52 | 2 | 2,11 | 128.3 | 2CH | |
| 123.22 | 2 | 12,1 | 123.21 | 2 | 12,1 | 125.5 | 4CH | |
| 123.08 | 2 | 14,3 | 123.08 | 2 | 10,3 | |||
| 118.27 | 2 | 10,5 | 118.26 | 2 | 14,5 | |||
| 66.44 | 2 | 7,8 | 66.43 | 2 | 7,8 | 62.8 | 2CH |
Dihydronaphthalene Oxide NMR Analysis
1R2S-Dihydronaphthalene Oxide 1S2R-Dihydronaphthalene Oxide
| 1R2S-Dihydronaphthalen | H-NMR svg | C-NMR svg | 1S2R-Dihydronaphthalen | H-NMR svg | C-NMR svg | Literature[5] | ||

|

|

|

|

|

|
H NMR (400 MHz, CDCl3) | C NMR (100 MHz, CDCl3) | |
| H-NMR | δ ppm | Degeneracy | Atom | δ ppm | Degeneracy | Atom | δ ppm | Multiplicity |
| 7.62 | 1 | 18 | 7.62 | 1 | 18 | 7.34 | d,J=7.2Hz,1H | |
| 7.39 | 2 | 20,19 | 7.39 | 2 | 20,19 | 7.09-7.25 | m, 2H | |
| 7.25 | 1 | 21 | 7.25 | 1 | 21 | 7.02 | d,J=7.2Hz,1H | |
| 3.56 | 1 | 6 | 3.56 | 1 | 6 | 3.79 | d,J=4.4Hz,1H | |
| 3.48 | 1 | 17 | 3.48 | 1 | 17 | 3.66 | t,J=4.0Hz,1H | |
| 2.95 | 1 | 15 | 2.95 | 1 | 16 | 2.64-2.70 | m,1H | |
| 2.27 | 1 | 16 | 2.27 | 1 | 15 | 2.47 | dd,J=5.6,15.6Hz,1H | |
| 2.21 | 1 | 7 | 2.21 | 1 | 8 | 2.29-2.37 | m,1H | |
| 1.87 | 1 | 8 | 1.87 | 1 | 7 | 1.62-1.72 | m,1H | |
| C NMR | 135.39 | 1 | 9 | 135.38 | 1 | 9 | 137.1 | |
| 130.37 | 1 | 10 | 130.37 | 1 | 10 | 132.9 | ||
| 126.67 | 1 | 11 | 126.67 | 1 | 11 | 129.9 | ||
| 123.79 | 1 | 13 | 123.79 | 1 | 13 | 128.83 | ||
| 123.53 | 1 | 14 | 123.53 | 1 | 14 | 128 | ||
| 121.74 | 1 | 12 | 121.74 | 1 | 12 | 126.5 | ||
| 52.82 | 1 | 2 | 52.82 | 1 | 2 | 55.5 | ||
| 52.19 | 1 | 1 | 52.19 | 1 | 1 | 55.2 |
Comment: NMR is not influenced by the enantiomers. And the value calculated is highly matched with the value in literature.
Optical Rotation
RR Stilbene SS Stilbene 1R,2S-Hydronaphthanlen 1S,2R-Hydronaphthanlen1 1S,2R-Hydronaphthanlen2
| Entry | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Epoxide | RR-Stilbene oxide | SS-Stilbene oxide | 1R,2S-Dihydronaphthalen oxide | 1S,2R-Dihydronaphthalen oxide (1) | 1S,2R-Dihydronaphthalen oxide (2) |
| Epoxide Structure | 
|
|
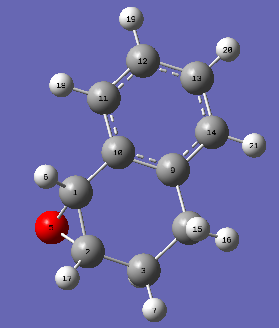
|
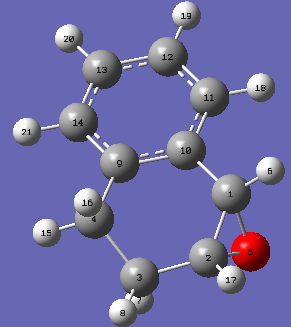
|
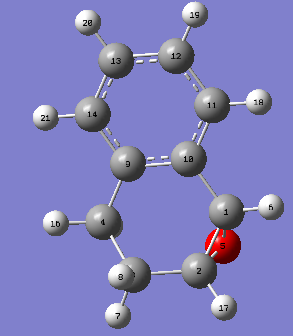
|
| Optical Rotation (589 nm)/deg | 297.70 | -298.22 | -35.86 | 35.86 | -155.82 |
| Literature Value | 250.8@22[6] | -249@15[7] | 129@20[8] | -39@25[9] |
All the optical rotation run in HPC under CAM-B3LYP/6-311++g(2df,p) and found in literature values are obtained in chloroform. The values for both RR/SS-stilbene oxide(+/-300 deg to +/- 250 deg) match very well with the literature value. However, for two dihydronaphthalene oxides, the calculated values match excellently well to each other, giving +/-35.86 deg. In comparison the literature values themselves have a big difference (129 deg/-36 deg). It may be cause by the favourable conformation of the dihydronaphthanlene is different in each cases. The entry 3 and 4 are generated by mirror image to each other and entry 5 is another conformation of 4. Entry 4 has H16 and H7 at the axial position, while entry 5 has H15 H8 at the axial position. Energy of entry 3 and 4 are exactly the same as -462.17535843 a.u. and entry 5 is -462.18120460 a.u.. They are almost the same in energy. It is likely the literature value of 129 deg is obtained while most the of 1S,2R-dihydronaphthanlene oxide is at a conformation of 5. If both of the literature values of dihydronaphthalene oxide were actually taken from the same conformation which is mirror image to each other, they should be same in value and opposite in direction as calculated.
Whether the sign is positive or negative doesn't predict its absolute configuration. In other word, it doesn't necessary mean whether it is R or S. It mainly depends on the molecule itself. Also by comparing entry 4 and 5, it shows that the conformation has a big impact on the optical rotation values as well as the sign. It may explain why a large variety of literature values have been reported.
Transition State
| Shi1 | Shi2 | Shi3 | Shi4 | Jacob | |
| Sum of electronic and thermal Free Energies(RR) | -1534.687808 | -1534.687252 | -1534.700037 | -1534.699901 | -3574.921174 |
| Sum of electronic and thermal Free Energies(SS) | -1534.683440 | -1534.685089 | -1534.693818 | -1534.691858 | -3574.923087 |
| Energy difference in (Hartree) | 0.004368 | 0.002163 | 0.006219 | 0.008043 | -0.001913 |
| Energy difference in (kJ/mol) | 11.468184 | 5.6789565 | 16.3279845 | 21.1168965 | -5.0225815 |
| Ratio of concentration of two species K | 102.3910684 | 9.896151054 | 728.0115043 | 5030.235748 | 0.131701249 |
| RR | 0.990327985 | 0.908224473 | 0.99862828 | 0.999801242 | 0.116374572 |
| SS | 0.009672015 | 0.091775527 | 0.00137172 | 0.000198758 | 0.883625428 |
| ee%(RR) | 98.07% | 81.64% | 99.73% | 99.96% | -76.73% |
The procedure of the table is generated from the free energy difference between two diasteromeric transition states, K conversion via the formula RR/SS=K=exp(ΔG/RT), RR=K/(K+1), SS=1-RR, ee%=RR-SS, where RR/SS is the ratio of two enantiomers of trans-stilbene oxide, R is gas constant, T is room temperature,ee% is enantiomeric excess=([major]-[minor])/([major]+[minor]).
As can be seen, Shi catalyses RR-stilbenes and the Jacobsen catalyst favours SS-Stilbenes. Reaction with a shi-catalyst gives a higher ee% value than Jacobsen. In literature[10] it is reported over 95% for RR-stilbene oxide.
In reality, it is most likely that the reaction undergoes transition state Shi-3 for both RR/SS-enantiomers, because the free energy is the lowest in both enantiomers. It has been proved to give over 99% ee% value. Transition State 'shi-2' is the most unlikely case, because it gives a higher energy barrier. Therefore it gives the least ee% value in the calculation.
The literature value for Jacobsen transtilbene enantiomeric excess can not be found as Jacobsen works better with cis olefins to give epoxides with highest ee's (ee:76.73% (SS)). But the calculation result tells us Jacobsen favours SS-stilbene oxide than RR.
NCI
| RR1 | RR2 | RR3 | RR4 | ||||||||||||
|
|
|
| ||||||||||||
| SS5 | SS6 | SS7 | SS8 | ||||||||||||
|
|
|
|
| Jacobsen-SS | Jacobsen-RR | ||||||
|
|
Comment In non-covalent interactions analysis, different colours indicate different interactions. Blue: very attractive;green:mildly attractive; yellow:mildly repulsive;red:strongly repulsive. Shi:Comparing the colourful rings, which means the bond forming site in the transition state, RR3-shi, RR4-shi can see a very obvious blue colour around, whereas, RR2-shi can rarely see a blue colour. In SS transition states, the attraction areas are smaller and more hard to see in contrast to RR transition states. Looking at the real NCI interaction (the green area), again, RR3-shi and RR4-shi have a more continuous interaction phases, while RR1-shi,RR2-shi are more likely broken into pieces. Similarly, more little pieces can be found in SS transition states than a smooth piece. Overall, it indicates RR is more favourable than SS and transition state RR3-shi is more favourable. It matches the calculated value above. More delocalised interactions are found in Jacobsen transition states. The difference of the NCI between the two is less obvious than the shi ones. However, if look carefully, more light blue attractive areas can be found in the SS transition state. It means the SS is more favoured but not strongly favoured. The consequence of these two transition state is the lower enantiomeric excess than Shi's catalyst.
QTAIM
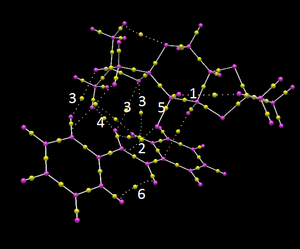

QTAIM analysis is complementary to the NCI analysis. Therefore RR3-shi is chosen to be analysed here. 1is the Hydrogen bonding between the hydrogen atom on methyl group and the oxygen atom at the C=O site, where the other oxygen forms the epoxide with the olefin. 5 is the Bond critical point, where shared electron density reaches a minimum. In other word, the bond is likely to break up here, due to the longer distance, weaker interaction between the oxygen atom and the BCP. As there is still a proper bond here, it indicates it is an early transition state. The transition state is alike to the starting material. 2 is the NCI between the oxygen and the olefin. It is close to one carbon. Also the interaction only appears at one of the epoxide carbon, not both. It could indicate C-O bond formation one after the other. This gives us a bit of idea how the mechanism goes as well. 3and4 are the through space interactions between the 5-membered ring with a chiral centre and the stilbene. Because most of the interactions of the transition state happen here, it mean it can only favour one side but not the other side. 3 is the hydrogen bond between the oxygen atom of the 5-membered ring and the hydrogen at the phenyl ring. There are three of them which contribute to the stabilisation of the transition state. 4 is the interaction between hydroden atoms. 6 is the interaction of hydrogen atoms at the closest approach.
As mentioned above, the possible mechanism can be suggested here.
New Candidate for Investigation
Optical Rotation: 64 deg, concentration:1.22 g/100ml, Solvent: Chloroform. Wavelength: 589 nm [12]
Molecular Weight: 138.21
This one is particularly interesting as the starting material has two C=C bonds. Therefore the regioselectivity in reaction works very well.
References
- ↑ W. F. Maier and P. v. R. Schleyer, J. Am. Chem. Soc., 1981, 103, 1891 DOI: [10.1021/ja00398a003]
- ↑ L. Paquette, N. A. Pegg, D. Toops, G. D. Maynard, R. D. Rogers, J. Am. Chem. Soc.,, 1990, 112, 277-283. DOI:10.1021/ja00157a043
- ↑ D. H. R. Barton J. Chem. Soc., 1948, 340-342 DOI: [10.1039/JR9480000340]
- ↑ L. Ji, Y. Wang,C. Qian,X. Chen,Synthetic Communications,2013,43:16,2256-2264 DOI: [10.1080/00397911.2012.699578]
- ↑ L. Ji, Y. Wang,C. Qian,X. Chen,Synthetic Communications,2013,43:16,2256-2264 DOI: [10.1080/00397911.2012.699578]
- ↑ D,J. Fox,D. S. Pedersen,A. B. Petersen,S. Warren, Org. Biomol. Chem., 2006, 4(16), 3117-3119 DOI: [10.1039/B606881B]
- ↑ J. Read, I. Campbell, J. Chem. Soc., 1930, 2377-2384 DOI: [10.1039/JR9300002377]
- ↑ Archelas; Furstoss; Pedragosa-Moreau Tetrahedron,1996,52,4593 - 4606 DOI: [10.1016/0040-4020(96)00135-4]
- ↑ H. Lin,Y. Liu,J. Qiao,Z Wu, Journal of Molecular Catalysis B: Enzymatic,2010,67,236 - 241 DOI: [10.1016/j.molcatb.2010.08.012]
- ↑ Y. Shi, Z. Wang, Y. Tu, M. Frohn, J. Zhang,J. Am. Chem. Soc., 1997, 119,, 11224-11235 DOI: [10.1021/ja972272g]
- ↑ S. S. Balula,N.Bion, S Bruno,A. C. Coelho, I. S. Goncalves, M. Pillinger,J. Rocha,A. Anabela,J. C. Alonso,P. Ferreira,N. Bion, Adv. Synth. Catal., 2010, 352,1759 - 1769 DOI: [10.1002/adsc.201000042]
- ↑ M. Kassiou,S. M. Wilkinson,J. Price,M. Kassiou, Tetrahedron Letters, 2013, 54, 52 - 54 DOI: [10.1016/j.tetlet.2012.10.080]