Rep:Mod:chenylbenzene
Haber-Bosch Process
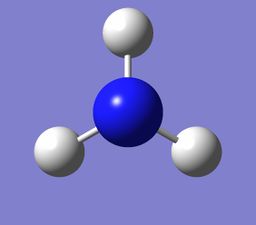
NH3
Ammonia |
Item Value Threshold Converged? Maximum Force 0.000006 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000014 0.001800 YES RMS Displacement 0.000009 0.001200 YES
The optimisation file is liked to here
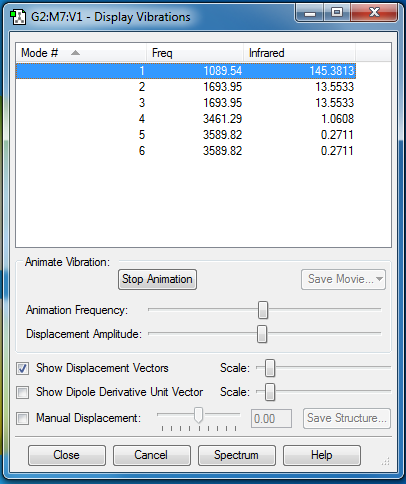
6 modes are expected from the 3N-6 rule.
Mode 2 and 3, and mode 5 and 6 are degenerate.
1-3 are "bending" vibrations and 4-6 are "bond stretch" vibrations.
Mode 1, 3, and 4 are highly symmetric.
Mode 1 is the "umbrella" mode.
6 bands are expected to be seen in an experimental spectrum of gaseous ammonia.
H2
Hydrogen |
| Information | ||
|---|---|---|

| ||
| A Gaussview image of an optimised H2 molecule. | ||
| More Info | ||
| Molecule Name | H2 | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Energy(au) | -1.17853936 | |
| Point Group | D∞h | |
| Bond Length | 0.74279 | |
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES
The optimisation file is liked to here
N2
Nitrogen |
| Information | ||
|---|---|---|

| ||
| A Gaussview image of an optimised N2 molecule. | ||
| More Info | ||
| Molecule Name | N2 | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Energy(au) | -109.52359111 | |
| Point Group | D∞h | |
| Bond Length | 1.09200 | |
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
The optimisation file is liked to here
Energy Change of Haber-Bosch Process
N2 + 3H2 -> 2NH3.
E(NH3) = -56.55664124 au
2*E(NH3) = -113.1132825 au
E(N2) = -109.52359111 au
E(H2) = -1.17853936 au
3*E(H2) = -3.53561808 au
ΔE = 2*E(NH3)-[E(N2)+3*E(H2)] = -0.05407331 au = -141.97 kJ/mol
Since the total energy change is negative, the product is more stable than the reactants.
Others
F2
Fluorine |
Item Value Threshold Converged? Maximum Force 0.000128 0.000450 YES RMS Force 0.000128 0.000300 YES Maximum Displacement 0.000156 0.001800 YES RMS Displacement 0.000221 0.001200 YES
The optimisation file is liked to here
The energy of bonding orbitals is lower than that of the atomic orbitals, and the energy of the antibonding orbitals is higher. Hence, bonding orbitals stabilize the molecule and antibonding orbitals destabilize it.
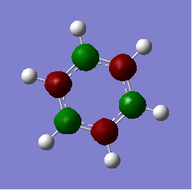
Benzene
Benzene |
Item Value Threshold Converged? Maximum Force 0.000198 0.000450 YES RMS Force 0.000082 0.000300 YES Maximum Displacement 0.000814 0.001800 YES RMS Displacement 0.000293 0.001200 YES
The optimisation file is liked to here
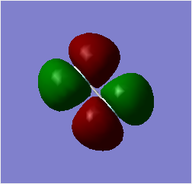
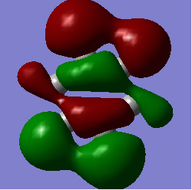
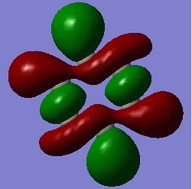
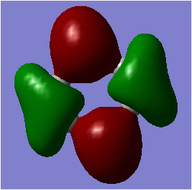
MO = 1 is the LCAO of all 1s orbitals, and this bonding orbital has the lowest energy (-10.18801).
MO = 21 (HOMO) is the LCAO of pz orbitals of all carbon atoms, and it is an antibonding orbital.
MO = 22 (LUMO) is the LCAO of pz orbitals of all carbon atoms as well, but this orbital has more nodes, and thus it has higher energy (0.00264) than the HOMO (-0.24690) does.