Rep:Mod:cheese
Mini Project: Assigning Regisomers in Click Chemistry
Introduction
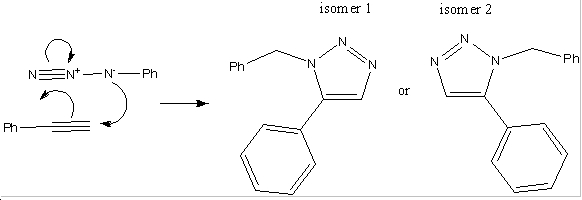
There have been developments made in the field of 1,3-dipolar cycloadditons between alkynes and azides. It has been shown that using an Cu(I) catalyst speeds up the reaction to such an extent that reactions of that kind are now referred to as click reactions. When subsititued azides and alkynes are used in click reactions, there are two possible regioisomers which can be formed. When the copper catalyst is used, it turns out that the 1,-4 isomer predominates, however when a ruthenium catalyst is used, this favours the production of the 1-5 isomer. Below is an example of such a reaction[1]:
Part 1
The aim of this section is to calculate the 13C spectra of the two isomers and to compare them with the literature values to see which shows the best comparison. This was achieved by first drawing and optimising the isomers in ChemBio3D using the MM2 calculation. The result of the initial optimisation was as follows:
Table 12: Initial Optimisation of Isomers 1 and 2 | |||||||
| Isomer 1 | Isomer 2 | ||||||
|
| ||||||
| 47.56 kcal/mol | 50.87 kcal/mol | ||||||
Then Gaussian input files were created, using the minimise function, and the method DFT-mp1pw91, with the basis set set to 6-31G(d,p). The file was saved and then edited in word pad so that the top line read as follows:
- mpw1pw91/6-31(d,p) opt(maxcycle=25)
The files was resaved and then submitted to the SCAN service, which was done by logging in to the SCAN website at https://scanweb.cc.imperial.ac.uk/uportal2/ and then submitting the files to Chemistry Lab 2 wall. Once the jobs were completed the formatted checkpoint files were downloaded. Again the Gaussian input files were edited in word pad so that the top line read:
- mpw1pw91/6-31(d,p) NMR scrf(cpcm,solvent=chloroform)
This file was then re-submitted to the SCAN service. Once this was complete the Gaussian Log file was downloaded and opened in Gaussian. Under the results tab, the NMR data was available. The carbon nucleus was selected and a reference value: TTMS HF/6-31G(d) GIAO.
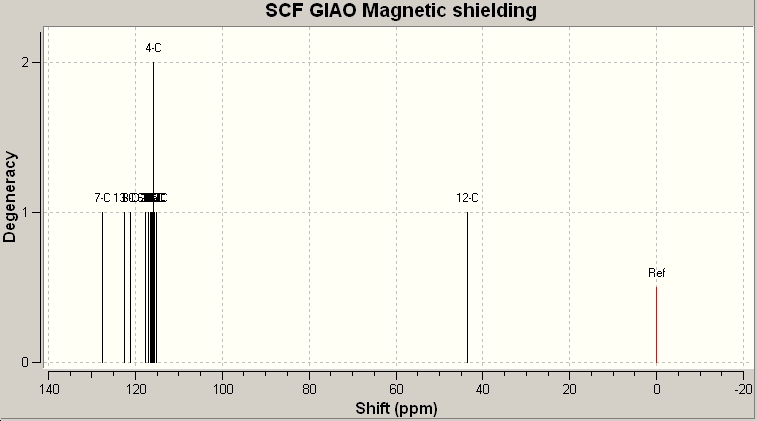
Isomer 1
Now we can compare the data calculated with that given by the literature:
Table 13: 13C NMR Data/ppm | |
| Calculated | Literature |
| 57.06 | 51.81 |
| 127.52 | 126.93 |
| 127.89 | 127.22 |
| 128.21 | 128.22 |
| 129.36 | 129.08 |
| 129.38 | 129.64 |
| 130.08 | 133.26 |
| 133.61 | 133.3 |
| 134.97 | 135.66 |
| 139.94 | 138.26 |
The calculation run by the SCAN service was published on D-Space and is accessible via the follwing link:
http://hdl.handle.net/10042/to-1735
As the above table shows, there is a very strong agreement with the calcualted data, and the experimentally determined data. The next ideal step would be to compare the data obtianed for isomer one with that of isomer 2. Unfortunately there were persistent errors when trying to retrieve the files, so I have linked the Gaussian checkpoint file below:
References
- ↑ J. Am. Chem. Soc. 2005, 127, 15998. DOI:10.1021/ja054114s