Rep:Mod:chchanmod1
Module One: Structure and Spectroscopy (Molecular Mechanics / Molecular Orbital Theory)
Introduction
Molecular structure and its spectroscopic characteristics can now be modelled accurately, which gives an insight about reactivity and selectivity of a molecule before conducting an experiment. Structural optimisation is often based on theories such as steric hindrance, inter/intra-molecular interactions, stereoelectronic interactions, while quantum mechanical approximation is used to estimate molecular orbitals and spectroscopic information.
Modelling using Molecular Mechanics
Molecular Mechanics
The Molecular Mechanics(MM) approach is based upon force field mechanics, hence a non-quantum mechanics method. The steric energy of a molecule is defined by the following five parameters:
- Bond stretches
- Bond angles
- Dihedral angles
- Intermolecular forces, including both Van der Waals repulsion forces and dipolar electrostatic attraction forces
- Hydrogen bonds, which usually is a zero term
Equation 1 - Force Field Mechanics Equation on calculating Steric Energy of a Molecule
The first three terms of Equation 1 are related to the strain effects, and are minimised to zero when the molecule is unstrained and unhindered. The non-bonded term can take on negative values, particularly if there are numerous inter/intra-molecular attractive terms or favourable ionic interactions. The Hydrogen bond term is considered as just between two specific atoms, thus no de-localised Hydrogen bonds are covered. π-conjugation and stereo-electronic (anomeric) effects are often being taken into account by the addition of two more terms in the sum. The main assumption of force field mechanics is the five parameters do not have any interactions among them, hence it would start to break down when unusual bondings are involved.
The two models that are being used to find the closest low energy conformation during this module are MM2 and MMFF94, as different models based on different force field formulae, thus the energies calculated can only be compared with similar molecules such as isomers and conformers, also energies calculated from different models cannot be compared.
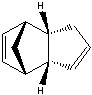
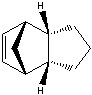
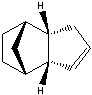
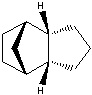
The Hydrogenation of Cyclopentadiene Dimer
|
The endo-dimer has a more favourable transition state due to more favourable orbital overlap in comparison with exo-dimer's transition state.  |
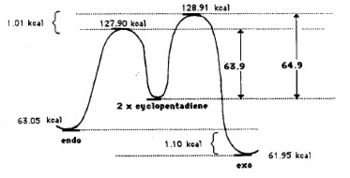 |
Product 1 and 2 are results of hydrogenation of one C=C bond of the endo-dimer, whereas product 3 is the result of both C=C bonds being reduced. The energy of product 2 is lower than product 1, which suggests formation of product 2 is thermodynamically more favourable. Both product 1 and 2 can undergo further hydrogenation to give product 3, but the reaction would not be thermodynamically favourable; this can be explained by the similar energies of product 1 and 2 and the energy of product 3 is higher than product 2. To make the reaction happens, nickel(II) chloride with water can be used to catalyse it, and the first C=C bond being reduced is the one as product 2 which agrees with the calculation [2]. Based upon the calculations, the key energy that distinguish between product 1 and 2 is bend energy. Product 1 has higher bend energy (~4 kcal/mol) than product 2, which mainly causes the destabilisation of product 1. Although product 3 has the lowest bend energy among the three, but it has the highest torsion energy and therefore causes destabilisation of the molecule.
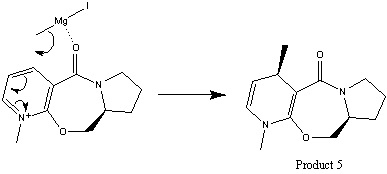
Stereochemistry of Nucleophilic Additions to a Pyridinium Ring (NAD+ Analogue)
The two reactions below demonstrate nuclephilic additions to pyridinium rings, both products have specific stereochemistry and this can be predicted based on the geometry optimised structures.
|
The second reaction is nucleophilic addition of NHPh group to pyridinium ring, unlike the above reaction, the oxygen does not coordinate with the nucleophile. The lowest energy conformation obtained by MM2 is with oxygen at the lower plane and therefore increases the steric hindrance of the lower plane. The attack of the nucleophile is at the upper plane so the steric effects can be minimised due to the bulky phenyl group of the nucleophile giving a stereochemically pure product. |
 |
Stereochemistry and Reactivity of an Intermediate in the Synthesis of Taxol
The synthesis of taxol involves an intermediate which demonstrates atropisomerism; compound 8 and 9 differ in the conformation of C=O bond, whether it is pointing up or down. Both compounds were minimised by MM2, the results show that compound 9 is more stable than compound 8, thus during the reaction compound 8 would convert to compound 9 to give a more stable intermediate can be expected. The major difference between the two compounds is their relative torsion energy due to the rotation of C=O bond. MMFF94 approach agrees with MM2, but since two methods have different combination of force field, hence the energies calculated are different.
| Compound 8 | Compound 9 | |
|---|---|---|
| Structure |  |

|
| 3-D Structure |
|
|
| MM2 Calculated Total Energy | 48.89 kcal/mol | 44.35 kcal/mol |
| MMFF94 Calculated Total Energy | 70.54 kcal/mol | 60.56 kcal/mol |
The alkane product of the intermediate is at a higher energy level (68.37 kcal/mol as estimated by MM2), this suggests that the C=C bond causes stabilisation of the compound. This is known as hyperstable alkene[3], which would occur when C=C bond adjacent to a bridgehead, hence the structure suffers less strain.
Modelling Using Semi-empirical Molecular Orbital Theory
MM approach is useful when optimising geometries of molecules and calculating their relative energies. As it is a non-quantum mechanics method, it cannot compute electrons, hence orbital interactions. Semi-empirical molecular orbital(MO) theory is introduced to take into account electron interactions, hence a quantum mechanics approach.
Regioselective Addition of Dichlorocarbene
Compound 10 was chosen to study the regioselectivity of dichlorocarbene addition, as there are two possible sites to perform the addition as shown in Figure 6.
|
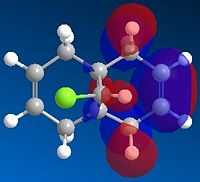
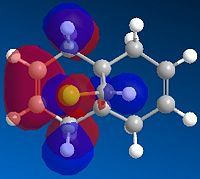
The geometry of the compound was minimised by MM2 to give a total energy of 17.00 kcal/mol. MOPAC/RM1 MO method was performed to generate the molecular orbitals of the structure, which can be used to distinguish between the two C=C bonds. The key molecular orbitals are shown in Table 5. |
 |
| HOMO-1 | HOMO | LUMO | LUMO+1 | LUMO+2 |
|---|---|---|---|---|
 |
 |
 |
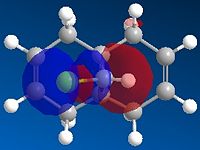 |
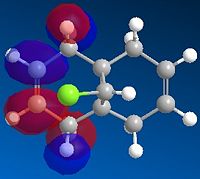
|
These molecular orbitals demonstrate an anti-periplanar stabilising overlap of the syn π orbital in the HOMO-1 and the empty σ* C-Cl of the LUMO +1, which lowers the energy of the syn C=C bond relative to the anti C=C bond. This results in syn C=C bond as a poorer electron acceptor, thus product 13 would be formed due to both stereoelectronic effects and steric effects.
Besides molecular orbital analysis, vibrational stretching can also be used to identify the site of addition. The infra-red spectrum of Compound 12 was calculated by Gaussian and the key peaks were reported in Table 6.
| Compound 12 | Compound 15 | |||
| 3-D Structure | ||||
| Wavenumber (cm-1) | Vibration Image | Wavenumber (cm-1) | Viration Image | |
| C-Cl Stretch | 770.86 | 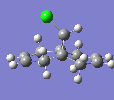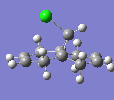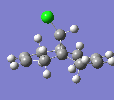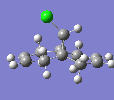 |
780.03 | 
|
| syn-periplanar C=C stretch | 1757.34 |  |
1753.72 | 
|
| anti-periplanar C=C stretch | 1737.03 |  |
- | |
 |
 |
|
The vibrational frequency of syn C=C bond is higher than that of anti C=C bond, which indicates anti C=C bond is weaker and is easier to break, hence it is more likely to be attacked by nucleophiles.
|
 |
Structure Based Mini Project[4] Using DFT-based Molecular Orbital Methods
Mini Project Introduction
Carbon-carbon bond forming reactions are crucial in organic synthesis and polymer chemistry, especially ring closing metathesis reaction, which plays an essential role. It is a powerful tool in synthesising macro-cyclic system, such as the key step in synthesising BILN 2061 ZW, which is a chemical entity showing promising results against the Hepatitis C virus.

The specific reaction chosen for this project is shown in Figure 11. The reaction conditions would give 84% as experimental yield and the mole ratio of cis- to trans-product is 2:3. The diaselectivity of the reaction is the result of the steric effects cause by the ligands around Ru atom.

Conformational Analysis
MM2 was performed to obtain the conformations with the minimum total energies for both cis- and trans- product. The results are shown in Table 7.
| Cis-product 25 | Trans-product 25 | |
|---|---|---|
| Structure |  |
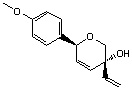
|
| 3-D Structure |
|
|
| Total Energy | 13.51 kcal/mol | 14.31 kcal/mol |
| Stretch Energy | 1.11 kcal/mol | 1.11 kcal/mol |
| Bend Energy | 4.62 kcal/mol | 4.44 kcal/mol |
| Stretch-Bend Energy | 0.20 kcal/mol | 0.22 kcal/mol |
| Torsion Energy | -3.06 kcal/mol | -2.92 kcal/mol |
| Non-1,4 Van der Waals Energy | -5.34 kcal/mol | -3.89 kcal/mol |
| 1,4 Van der Waals Energy | 14.90 kcal/mol | 13.82 kcal/mol |
| Dipole-dipole Interaction Energy | 1.08 kcal/mol | 1.53 kcal/mol |
MM2 calculations show that cis-product is of a lower energy (~ 1 kcal/mol) than trans-product,hence thermodynamically more stable due to the stabilisation by torsion energy, non-1,4 Van der Waals energy and dipole-dipole interaction energy. The synthetic experiment shows that trans-product has a higher mole ratio in the product mixture, this suggests that the reaction is kinetically governed, thus formation of trans-product goes via a more stable transition state with smaller activation energy barrier.
13C NMR Spectrum Estimation
After minising the structures by MOPAC/RM 1 MO method, the data were processed by Gaussian to give the 13C NMR estimations.
|
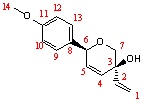 |
Based upon Table 8, the Carbon-10 is not observed in the experimental NMR data since it is in the same environment as Carbon-12, hence one peak with double intensity would be shown. The same principle applies to Carbon-9 and -13.
|
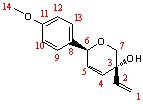 |
In comparison with the literature of Trans-product, Carbon-9 and -12 should be merged with Carbon-13 and-10 respectively due to two identical Carbon environments; this can explain the relatively large deviations for those peaks.
The key peak in identifying the two isomers is Carbon-7 as in the literature they have the largest difference comparing with relative Carbon environment; this effect is arisen by the chiral centre (Carbon-3).
Conclusion
Based upon the results obtained 13C NMR is not the best method in identifying the two products, as most peaks are very similar. Alternatively 1H NMR would be a better spectroscopic approach, as the chemical shifts and coupling constants are different between the two products.
Reference
- ↑ M. A. Fox, R. Cardona and N. J. Kiwiet, Journals of Organic Chemistry, 1987,52, 1469-1474
- ↑ F. Alonso and M. Yus, Tetrahedron Letters, 1996, 37, 6925-6928
- ↑ W. Maier and P. Schleyer, J. Am. Chem. Soc., 1981, 103, 1891-1893
- ↑ B. Schmidt and M. Westhus, Tetrahedron, 2000, 56, 2421-2426
- ↑ T. Nicola, M. Brenner, K. Donsbach and P. Kreye, Organic Process Research & Development, 2005, 9, 513-515