Rep:Mod:ch6518y1imm2
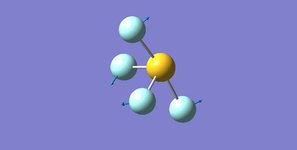
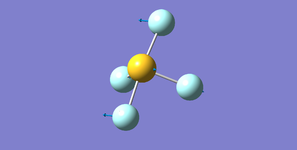
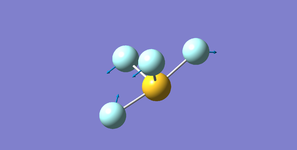
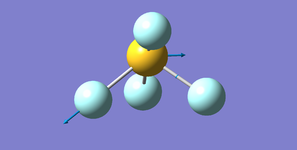
Ammonia NH3
Structure
| Calculation Method | Basis Set | Final Energy E / a.u. | Point Group |
|---|---|---|---|
| RB3LYP | 631G(d,p) | -56.55776873 | C3V |
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES
NH3 |
Optimised N-H bond length (Angstrom): 1.02 ± 0.01
Optimised H-N-H bond angle (Degrees): 106 ± 1
Charge on N: +0.375
Charge on H: -1.125
(Dipole Moment: 1.8466 Db)
Opt file here
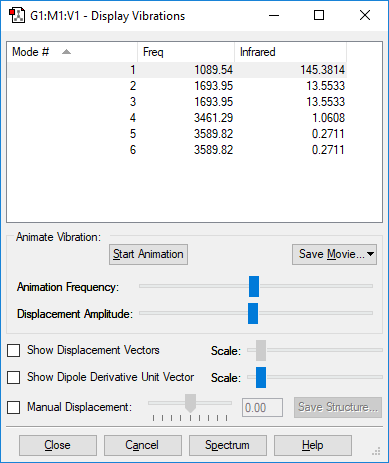
Vibrational Modes
| Wavenumber / cm-1 | Symmetry | Intensity | Bend or Stretch | Image |
|---|---|---|---|---|
| 1090 | A1 | 145.4 | Bend | 
|
| 1694 | E | 13.6 | 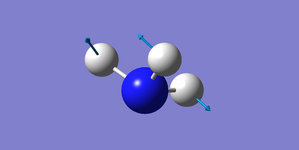
| |
| 1694 | E | 13.6 | 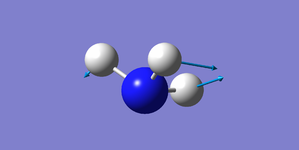
| |
| 3461 | A1 | 1.1 | Stretch | 
|
| 3590 | E | 0.3 | 
| |
| 3590 | E | 0.3 | 
|
Number of expected vibrational modes:
Two pairs of degenerate vibrational modes, one pair at 1694 cm-1, the other at 3590 cm-1.
The vibrations at 1090 cm-1 and 3461 cm-1 are highly symmetric.
The 1090 cm-1 is known as the "umbrella" mode due to its resemblance to an opening and closing umbrella.
In an IR spectrum of gaseous ammonia, four bands would be predicted since there are vibrational modes at four different frequencies.
Haber-Bosch Process
Molecular Nitrogen N2
Structure
| Calculation Method | Basis Set | Final Energy E / a.u. | Point Group |
|---|---|---|---|
| RB3LYP | 631G(d,p) | -109.52412868 | D∞h |
Item Value Threshold Converged? Maximum Force 0.000001 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES
N2 |
Optimised N≡N bond length (Angstrom): 1.11 ± 0.01
Charge on each N: 0
(Dipole moment: 0 Db)
The molecule (dinitrogen)-(2,2',2' '-(phosphanetriyl)tris(1-(diphenylphosphanyl)-3-methyl-1H-indole))-ruthenium tetrahydrofuran solvate (refcode DEKFUX) has a reported N≡N bond length of 1.086(6) Angstrom. In dinitrogen complexes, one of the nitrogens donates its lone pair, which exists in an sp orbital, to the metal to form a dative covalent bond. Consequently, the electron density around the donor nitrogen decreases, causing it to attract the electrons in the N≡N pi-bond more strongly since the effective nuclear charge they experience increases. Because the electrons move closer to the bonded nitrogen, so too does the second nitrogen; this results in a shorter equilibrium bond length. In pure diatomic molecular nitrogen gas, both atoms are electronically equal, but when coordinated with a metal there is an imbalance in electron density.
Opt file here
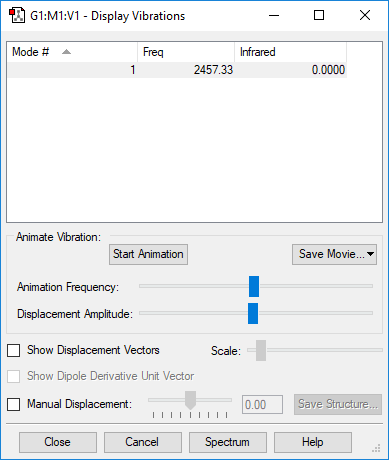
Vibrational Modes
| Wavenumber / cm-1 | Symmetry | Intensity | Bend or Stretch | Image |
|---|---|---|---|---|
| 2457 | SGG | 0.0 (IR inactive) | Stretch | 
|
Number of expected vibrational modes:
Molecular Hydrogen H2
Structure
| Calculation Method | Basis Set | Final Energy E / a.u. | Point Group |
|---|---|---|---|
| RB3LYP | 631G(d,p) | -1.17853936 | D∞h |
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES
H2 |
Optimised H-H bond length (Angstrom): 0.74 ± 0.01
Charge on each H: 0
(Dipole moment: 0 Db)
Opt file here
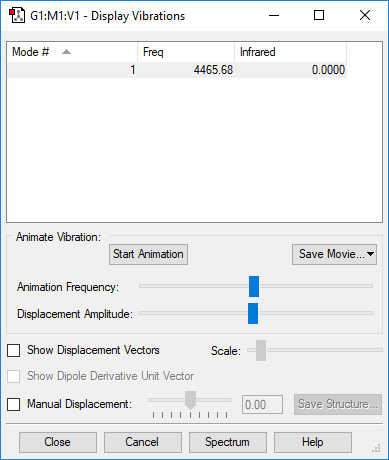
Vibrational Modes
| Wavenumber / cm-1 | Symmetry | Intensity | Bend or Stretch | Image |
|---|---|---|---|---|
| 4466 | SGG | 0.0 (IR Inactive) | Stretch | 
|
Number of expected vibrational modes:
Energy Calculations
Hence ΔE = -146.5 kJ mol-1 < 0
So the reaction is exothermic, and thus the ammonia product is more stable than the reactants. However, entropy greatly favours the reverse reaction.
Sulfur Tetrafluoride SF4
Structure
| Calculation Method | Basis Set | Final Energy E / a.u. | Point Group |
|---|---|---|---|
| RB3LYP | 631G(d,p) | -797.45952460 | C2v |
Item Value Threshold Converged? Maximum Force 0.000148 0.000450 YES RMS Force 0.000065 0.000300 YES Maximum Displacement 0.000570 0.001800 YES RMS Displacement 0.000281 0.001200 YES
SF4 |
Optimised S-Fax bond length (Angstrom): 1.67 ± 0.01
Optimised S-Feq bond length (Angstrom): 1.59 ± 0.01
Optimised Fax-S-Fax bond angle (Degrees): 171 ± 1
Optimised Feq-S-Feq bond angle (Degrees): 102 ± 1
Optimised Fax-S-Feq bond angle (Degrees): 87 ± 1
Charge on S: +1.983
Charge on Fax: -0.537
Charge on Feq: -0.455
(Dipole moment: 0.8849 Db)
Opt file here
Vibrational Modes
Number of expected vibrational modes:
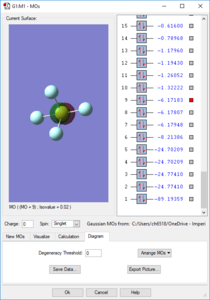
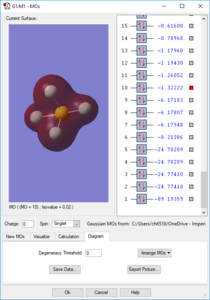
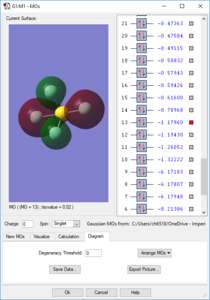
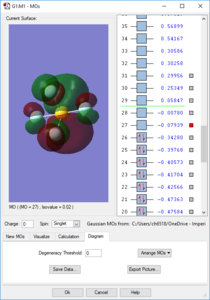
Molecular Orbitals
Orbitals 1-9 are very deep in energy and are therefore inaccessible; these are made from the core electrons of the constituent atoms. However, subsequent MOs are much higher-lying, with all MOs above the 14th having E > -1 eV. Overall the molecule has no pi bonds, therefore for every filled bonding orbital with pi character there must be an equal amount of pi antibonding character. Due to the number of atomic orbitals contributing to the molecular orbitals, none of the higher-lying orbitals can be considered to be purely bonding or purely antibonding. The electron distribution for the orbitals may lie between one pair of atoms, but produce a node between another pair. S-character can be observed in the shape of orbital 14, whilst the large rear lobe(s) of orbitals 26 and 27 (the HOMO and LUMO) indicate contribution from a d orbital.
Independence Task - other small molecules
Carbon Monoxide CO
Structure
| Calculation Method | Basis Set | Final Energy E / a.u. | Point Group |
|---|---|---|---|
| RB3LYP | 631G(d,p) | -113.30945314 | C∞v |
Item Value Threshold Converged? Maximum Force 0.000007 0.000450 YES RMS Force 0.000007 0.000300 YES Maximum Displacement 0.000003 0.001800 YES RMS Displacement 0.000004 0.001200 YES
CO |
Optimised C-O bond length (Angstrom): 1.14 ± 0.01
Charge on C: +0.506
Charge on O: -0.506
(Dipole moment: 0.0599 Db)
Opt file here
Vibrational Modes
| Wavenumber / cm-1 | Symmetry | Intensity | Bend or Stretch | Image |
|---|---|---|---|---|
| 2209 | SG | 68.0 | Stretch | 
|
Number of expected vibrational modes:
Cyanide CN-
Structure
| Calculation Method | Basis Set | Final Energy E / a.u. | Point Group |
|---|---|---|---|
| RB3LYP | 631G(d,p) | -92.82453153 | C∞v |
Item Value Threshold Converged? Maximum Force 0.000012 0.000450 YES RMS Force 0.000012 0.000300 YES Maximum Displacement 0.000005 0.001800 YES RMS Displacement 0.000008 0.001200 YES
CN- |
Optimised C-N bond length (Angstrom): 1.18 ± 0.01
Charge on C: -0.246
Charge on N: -0.754
(Dipole moment: 0.5236 Db)
Opt file here
Vibrational Modes
| Wavenumber / cm-1 | Symmetry | Intensity | Bend or Stretch | Image |
|---|---|---|---|---|
| 2139 | SG | 7.8 | Stretch | 
|
Number of expected vibrational modes:
Hydrogen Cyanide HCN
Structure
| Calculation Method | Basis Set | Final Energy E / a.u. | Point Group |
|---|---|---|---|
| RB3LYP | 631G(d,p) | -93.42458132 | C∞v |
Item Value Threshold Converged? Maximum Force 0.000370 0.000450 YES RMS Force 0.000255 0.000300 YES Maximum Displacement 0.000676 0.001800 YES RMS Displacement 0.000427 0.001200 YES
HCN |
Optimised C-N bond length (Angstrom): 1.07 ± 0.01
Optimised H-C bond length (Angstrom): 1.16 ± 0.01
Charge on H: +0.234
Charge on C: +0.073
Charge on N: -0.308
(Dipole moment: 2.8933 Db)
Opt file here
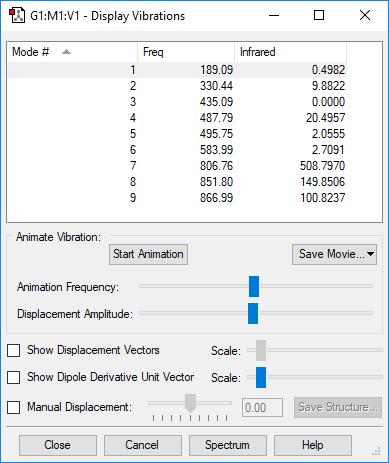
Vibrational Modes
| Wavenumber / cm-1 | Symmetry | Intensity | Bend or Stretch | Image |
|---|---|---|---|---|
| 767 | PI | 35.3 | Bend | 
|
| 767 | PI | 35.3 | 
| |
| 2215 | SG | 2.0 | Stretch | 
|
| 3480 | SG | 57.3 | 
|
Number of expected vibrational modes:
Pair of degenerate vibrational modes at 767 cm-1
Marking
Note: All grades and comments are provisional and subject to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have received your grade from blackboard.
Wiki structure and presentation 1/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES
NH3 0.5/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES, most answers are correct. However there are only 2 visible peaks in the spectra of NH3, due to the low intensity of the other 2 peaks. (See infrared column in vibrations table.)
N2 and H2 0.5/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES
Haber-Bosch reaction energy calculation 1/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 3/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
YES, you added all the necessary information, well done! To improve you could have explain the charges in terms of electronegativity, and added more details on the individual MOs such as which AOs are contributing to each. The explanation of the MOs overall was good though.
Independence 1/1
If you have finished everything else and have spare time in the lab you could:
Check one of your results against the literature, or
Do an extra calculation on another small molecule, or
Do some deeper analysis on your results so far
You did several extra calculations, with analysis, well done!