Rep:Mod:catherinelu
Introduction
Computational chemistry can be used to analyse molecules that cannot be characterised experimentally. It is a powerful tool in locating transition states, obtaining thermodynamic and kinetic data as well as predicting IR and NMR spectra, dipole moments and atom interactions.
In this study, GaussView 5.0 is used to create and analyse inorganic molecules using various methods and basis sets. Where more computational power was required, GaussView was used to create input files for submission to the HPC. Infrared spectra of molecules were predicted and MOs and NBOs were investigated to analyse bonding interactions.
Borane Optimisation
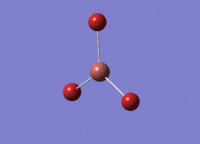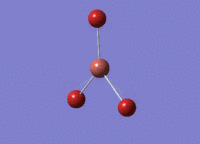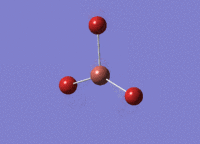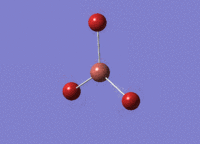
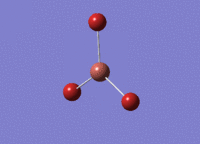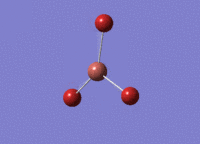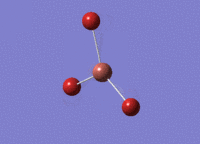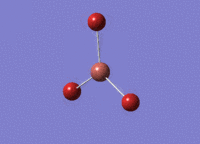
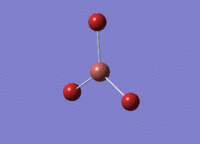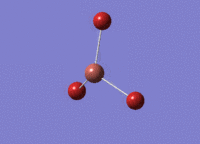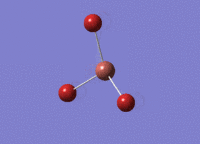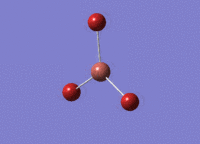
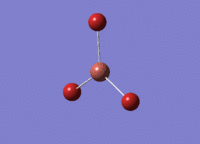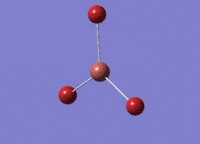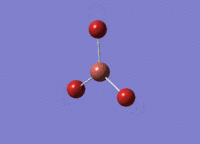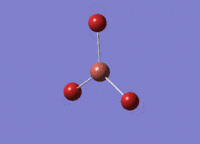
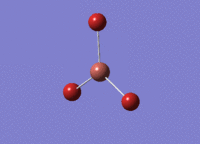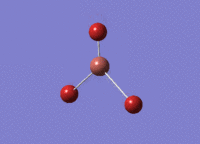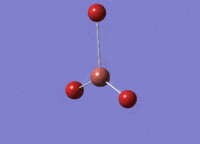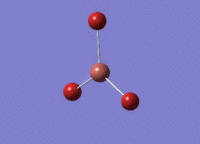
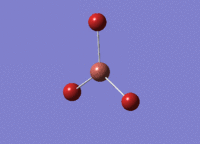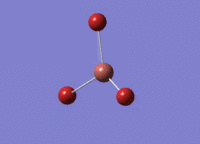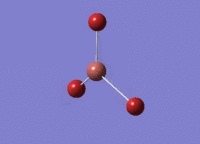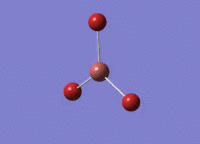
BH3 was drawn with trigonal planar geometry and bond lengths of 1.5Å. The B3LYP method and 3-21G basis set was used to optimise the molecule. Being a simple, symmetrical molecule, this method was sufficient.
Optimisation with method B3YLP and a 3-21G basis set produced the following .log file : File:BH3 OPT2CL.LOG
The optimisation process was successful and all parameters converged.
Item Value Threshold Converged?
Maximum Force 0.000413 0.000450 YES
RMS Force 0.000271 0.000300 YES
Maximum Displacement 0.001610 0.001800 YES
RMS Displacement 0.001054 0.001200 YES
Predicted change in Energy=-1.071764D-06
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1935 -DE/DX = 0.0004 !
! R2 R(1,3) 1.1935 -DE/DX = 0.0004 !
! R3 R(1,4) 1.1935 -DE/DX = 0.0004 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Optimisation Procedure
When a molecule is optimised, the Schrodinger equation is solved for varying positions of the nuclei (under the Born-Oppenheimer approximation, which states that the nuclei and electrons' positions are indenpendent of each other as the electrons are far less massive than the nuclei, and can adjust their positions instantaneously when the nuclei positions change).
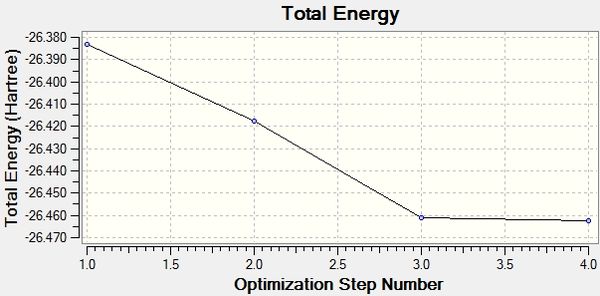
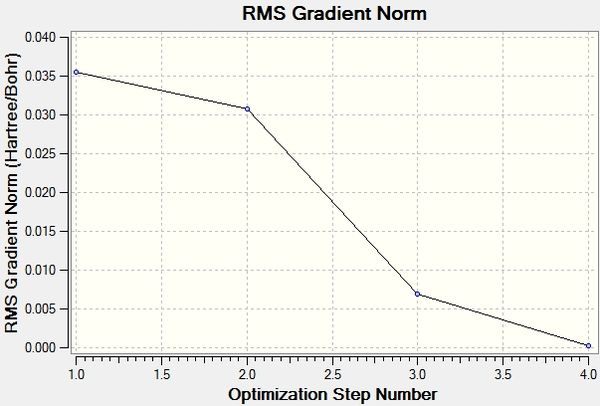
The Total Energy curve shows Gaussview traversing the potential energy surface to find structure of borane that corresponds to an energy minimum; each point on the graph corresponds to a combination of nuclei positions which increasingly resembles the stable structure. This occurs with several iterations until a point is found at which the gradient of the energy surface is equal to 0, i.e. a minimum. At this minimum, the short-range repulsion of the nuclei is in equilibrium with the long-range attraction of the nuclei with the electrons. A gradient is the change in x with respect to the change in y, dy/dx. Here, the y axis is the Energy and the x axis is the nuclei positions, so the gradient is dE/dx. The Root Mean Square Gradient Gradient shows the gradient approaching zero as Gaussview gets closer to the energy minimum. The final structure is that with the lowest energy and corresponds to the smallest RMS gradient.
Some of the intermediate structures shown by Gaussview may have no bonds. This is because Gaussview assigns bonds on a distance-based criteria. A bond is an attractive electrostatic interaction between two atoms' nuclei and electrons - therefore there may still be some degree of interaction and bonding in these structures that are not shown in Gaussview.
6-31G(d,p) Optimisation
The borane molecule optimised above was then run with a higher basis set of 6-31G(d,p) and a method of B3YLP: File:BH3 OPT 631G DPCL.LOG. The higher quality basis set enables a more accurate result. However, these results cannot be compared with those obtained above - results can only be compared when using the same basis set and method for all calculations.
Item Value Threshold Converged?
Maximum Force 0.000433 0.000450 YES
RMS Force 0.000284 0.000300 YES
Maximum Displacement 0.001702 0.001800 YES
RMS Displacement 0.001114 0.001200 YES
Predicted change in Energy=-1.189019D-06
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1914 -DE/DX = 0.0004 !
! R2 R(1,3) 1.1914 -DE/DX = 0.0004 !
! R3 R(1,4) 1.1914 -DE/DX = 0.0004 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
TlBr3 Optimisation
TlBr3 was optimised with a medium level basis set. With 81 electrons in Tl and 35 electrons in each Br, TlBr3 displays relativistic effects - these are not recovered in the standard Schrodinger equation. Therefore a pseudo-potential was used to help account for this, which makes the approximation that only the valence electrons contribute to bonding interactions. As Tl is a highly toxic element, another advantage of computational chemistry is exposed here - its properties may be investigated with no risk to the individual.
The completed optimisation was published in D-space [1].
The literature value found experimentally for the Tl-Br bond length of 2.512Å [1] is close to that calculated above, again showing the accuracy and power of computational chemistry.
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000014 0.001200 YES
Predicted change in Energy=-6.084108D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.651 -DE/DX = 0.0 !
! R2 R(1,3) 2.651 -DE/DX = 0.0 !
! R3 R(1,4) 2.651 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
BBr3 Optimisation
The completed optimisation was published in D-space [2].
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000036 0.001800 YES
RMS Displacement 0.000023 0.001200 YES
Predicted change in Energy=-4.027374D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.934 -DE/DX = 0.0 !
! R2 R(1,3) 1.934 -DE/DX = 0.0 !
! R3 R(1,4) 1.934 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Structure Comparison
| Molecule | Bond Distance (a.u.) |
|---|---|
| BH3 | 1.19349 |
| BBr3 | 1.93396 |
| TlBr3 | 2.65095 |
| Atom | Electron Configuration |
|---|---|
| B | [He] 2s2 2p1 |
| Br | [Ar] 4s2 3d10 4p5 |
| H | 1s1 |
| Tl | [Xe] 4f14 5d10 6s2 6p1 |
Both Boron and Thallium are in group 3 but B is in period 2 whereas Tl is in period 6. Tl therefore has much larger more diffuse valence orbitals and consequently poorer orbital overlap with Br than B has with H. Tl will have far more shielding of the nuclear charge by its numerous core electrons relative to B.
BBr3 bonding involves the sharing of an electron in Br’s 4p orbital with B’s 2p electron. BH3 bonding involves the sharing of H’s 1s electron and B’s 2p electron. By comparing BH3 and BBr3, it can be seen that BBr3 has a greater bond length. Br in period 4 has a greater atomic radius and more diffuse orbitals than H in period 1 and thus poorer orbital overlap with B. The Br ligands bonded to B experience a weaker effective nuclear charge as the Br nucleus’ positive charge is shielded by more core electrons than that of H, which has only one electron and thus experiences no shielding effect.
Both Br and H are non-metals with electronegativities of 2.96 and 2.20 respectively (Pauling Scale), compared to B with 2.04. The bonding character in BH3 will be covalent with comparatively little polarity in the B-H bonds, compared to BBr3 which is also covalently bonded but with a greater polarity in the B-Br bonds.
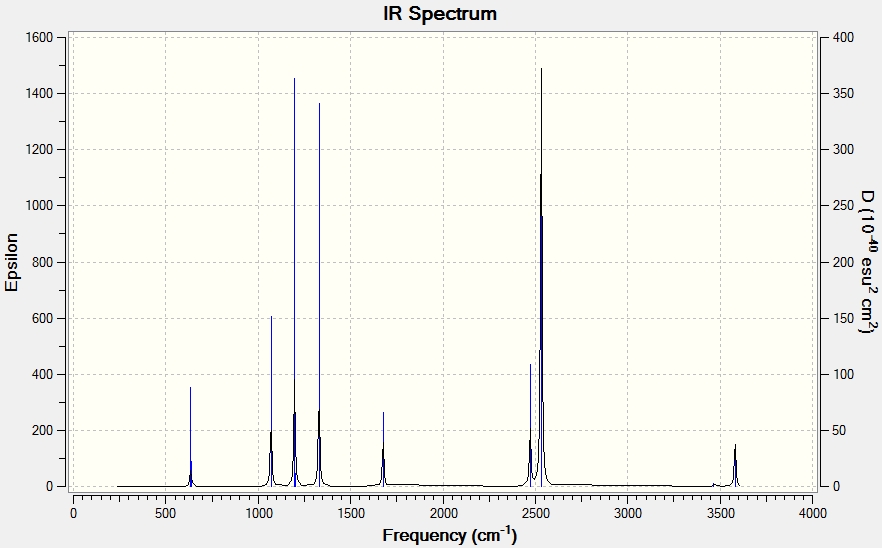
BH3 Frequency Analysis
The predicted bond length compares favourably to that of literature of 1.226 Å [2], suggesting that the optimisation has been successfully executed, as shown in the following:
Item Value Threshold Converged?
Maximum Force 0.000003 0.000015 YES
RMS Force 0.000002 0.000010 YES
Maximum Displacement 0.000013 0.000060 YES
RMS Displacement 0.000008 0.000040 YES
Predicted change in Energy=-6.170060D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1923 -DE/DX = 0.0 !
! R2 R(1,3) 1.1923 -DE/DX = 0.0 !
! R3 R(1,4) 1.1923 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
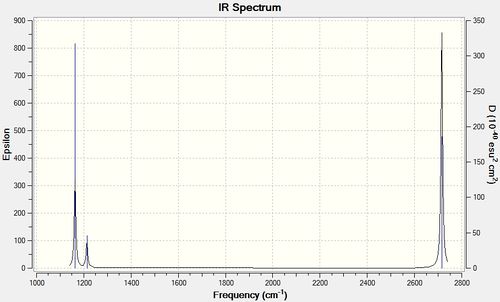
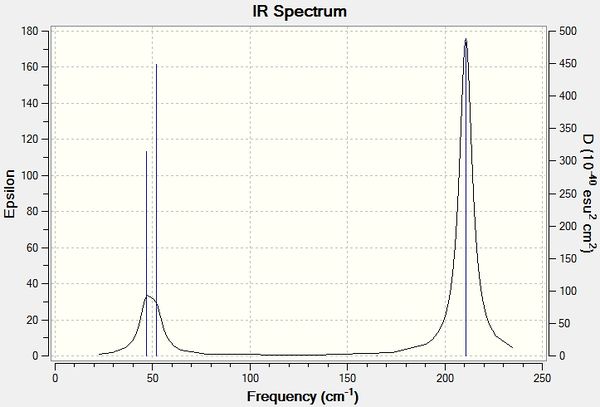
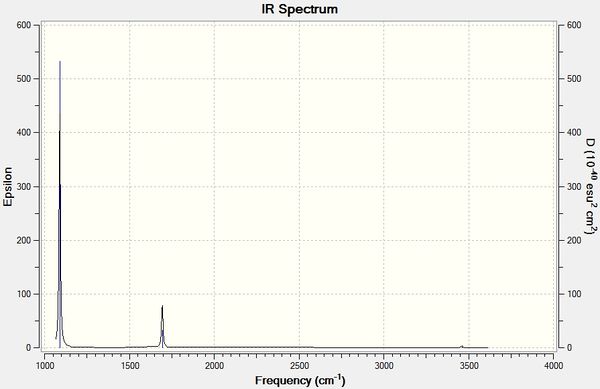
Low frequencies --- -7.0794 -7.0439 -0.0279 -0.0006 0.7084 6.6303 Low frequencies --- 1163.0023 1213.1577 1213.1579
Vibrational Analysis
The predicted spectrum only shows three peaks despite there being six vibrational modes. Modes 1 and 2 are degenerate and therefore occur at the same wavenumber, as are modes 5 and 6. Mode 4 has an IR intensity of 0.0000 and does not cause a change in the dipole moment as the stretch is symmetrical; this mode is therefore not infrared active and does not appear in the spectrum.
TlBr3 Frequency Analysis
The completed optimisation was published in D-space [3].
Item Value Threshold Converged?
Maximum Force 0.000001 0.000015 YES
RMS Force 0.000001 0.000010 YES
Maximum Displacement 0.000016 0.000060 YES
RMS Displacement 0.000011 0.000040 YES
Predicted change in Energy=-3.309373D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.6512 -DE/DX = 0.0 !
! R2 R(1,3) 2.6512 -DE/DX = 0.0 !
! R3 R(1,4) 2.6512 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Low frequencies --- -0.0013 -0.0006 -0.0001 1.8492 2.6414 2.6414 Low frequencies --- 46.6990 46.6991 51.9486
Vibrational Analysis
The lowest 'real' normal mode is 46.6990 cm-1.
Comparison of TlBr3 and BH3 Vibrational Frequencies
| Vibrational Mode | BH3 Frequency (cm-1) | TlBr3 Frequency (cm-1) |
|---|---|---|
| 1 | 1163 | 47 |
| 2 | 1213 | 47 |
| 3 | 1213 | 52 |
| 4 | 2582 | 165 |
| 5 | 2716 | 211 |
| 6 | 2716 | 211 |
The stretching absorption frequency of a bond can be approximated with Hooke's Law:
where µ is the reduced mass, f is the force constant of the bond and c is the velocity of light. Vibrational frequency depends on the bond strength and also the reduced mass; vibrational frequency is proportional to bond energy and inversely proportional to reduced mass.
It can be seen that BH3 vibrational frequencies are at much greater wavenumbers than that of TlBr3. This observation can be explained by Tl and Br being much heavier atoms, with atomic masses of 204.38 and 79.904 respectively, than B and H, with atomic masses of 10.81 and 1.008 respectively. The reduced masses of the vibrational modes of TlBr3 are significantly greater than those of BH3. The B-H bond is stronger than the Tl-Br bond as Tl and Br have larger, more diffuse orbitals and poorer orbital overlap. The Tl-Br bond length was seen to be longer than the B-H bond from the optimisations, showing the relative weakness of the Tl-Br bond.
Both spectra display three peaks; both of these have 2 peaks each which are from 2 degenerate vibrational modes. Both molecules have two pairs of degenerate E' vibrational modes, one A2" vibrational mode and one infrared inactive A1' vibrational mode. For both spectra the A2" and E' modes lie fairly closely together and the other two modes, A1' and E, also lie fairly close together and higher in energy. However, there is a different order of the A2" and E' modes for the two molecules - in BH3 the A2" mode is lower than the E' modes but in TlBr3 the A2" mode is higher than the E' mode. This reordering of the modes may be due to the mass of the Br and H. Br has a far greater mass than H; a concerted in and out of plane symmetrical bend (A2") takes far more energy than rocking from side to side in the plane, and so for TlBr3, the A2" is higher in energy than the E'. The converse is true for BH3; H has an atomic mass of 1 and the A2" symmetrical bend is lower in energy than the rocking side to side.
The same method and basis set is used for both the optimisation and frequency analysis calculations beacause the total energy is highly dependent which basis set is used and its quality; in order to be able to compare the energies of two molecules such as BH3 and TlBr3, they must have the same number of atoms and have used the same basis-set on every atom.
The low frequencies represent the motions of the molecule's centre of mass - every molecule has 3N-6 vibrational frequencies where N is the number of atoms; the low frequencies represent the '-6' vibrational frequencies. The positive values on the line after this represent the 'real' vibrational frequencies.
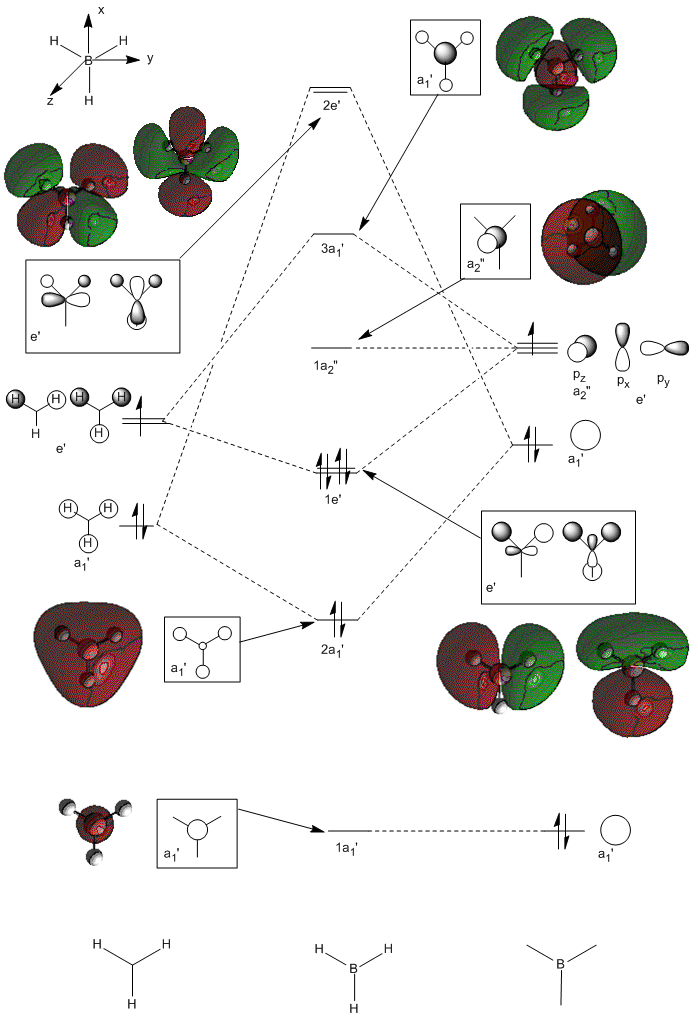
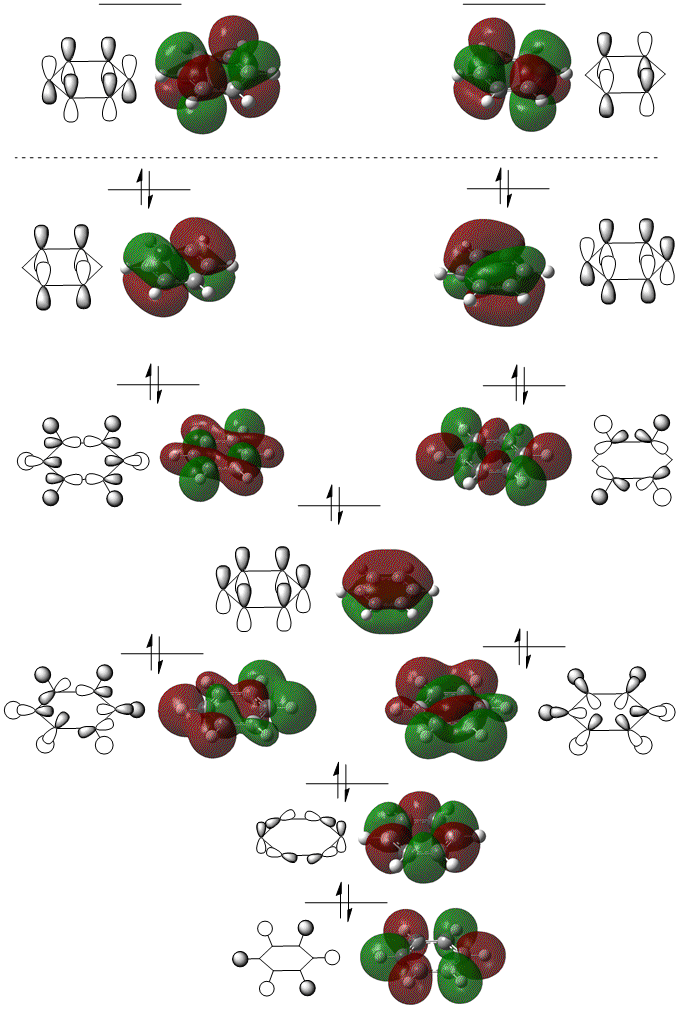
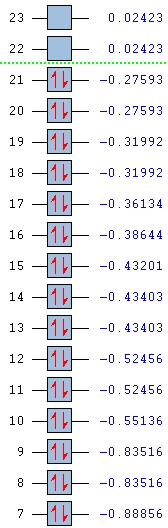
Molecular Orbitals of BH3
A population analysis was run of BH3 using the method as Energy and the basis set as 63G1(d,p). This was deposited in D-space: [4]
Orbital energies and kinetic energies (alpha):
1 2
1 (A1')--O -6.771122 10.797475
2 (A1')--O -0.512674 0.905366
3 (E')--O -0.350883 0.728735
4 (E')--O -0.350883 0.728735
5 (A2")--V -0.066018 0.640307
6 (A1')--V 0.168777 0.934518
7 (E')--V 0.179500 0.644102
8 (E')--V 0.179500 0.644102
The molecular orbitals were visualised and an MO diagram was drawn, showing the relative energies of the MOs. It can be seen that for this calculation and basis set, the degenerate 2e' orbitals are of higher orbital energy than the 3a1'.The fragment orbitals used were H3 and a central B atom.
For the H3 fragment, the lowest energy totally bonding orbital must be totally symmetric and of a1' symmetry. There are then two degenerate higher energy fragment orbitals of e' symmetry.
For the central B atom, the lowest energy atomic orbital is the s orbital, of a1' symmetry. The px and py orbitals are a degenerate pair of e' symmetry; the pz orbital is a2".
Boron and hydrogen are similar in electronegativity and so their s atomic orbitals are close in energy. The two a1' symmetry orbitals combine with very large splitting, as do the e' orbitals with somewhat smaller splitting. There are no H3 orbitals of a2" symmetry so this orbital remains non-bonding. This produces the following diagram:
The real MOs and those produced by LCAO do not differ significantly, showing the power and accuracy of qualitative MO theory.
NH3 NBO Analysis
Optimisation
The ammonia was initially optimised: File:NH3OPTCLU.LOG
Item Value Threshold Converged?
Maximum Force 0.000024 0.000450 YES
RMS Force 0.000012 0.000300 YES
Maximum Displacement 0.000079 0.001800 YES
RMS Displacement 0.000053 0.001200 YES
Predicted change in Energy=-1.629731D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7413 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7486 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7479 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8631 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Frequency
The frequency analysis was then performed: File:NH3FREQCLU.LOG
Item Value Threshold Converged?
Maximum Force 0.000004 0.000015 YES
RMS Force 0.000002 0.000010 YES
Maximum Displacement 0.000007 0.000060 YES
RMS Displacement 0.000004 0.000040 YES
Predicted change in Energy=-1.902561D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7447 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7444 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7443 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8637 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
The low frequencies are within a narrow range and all the 'real' low frequencies are positive.
Low frequencies --- -9.3571 -8.2143 -6.2890 -0.0017 -0.0012 -0.0007 Low frequencies --- 1089.3350 1693.9208 1693.9246
Modes 5 and 6 cause a very weak change in dipole moment and are not of great enough intensity to be observable in the infrared spectrum. Mode 1 causes a very significant change in dipole moment and thus causes the most intense peak.
Population
A population analysis was then carried out: File:NH3ENERGYCLU.LOG
| MO | Image | Orbital Energy | MO | Image | Orbital Energy |
|---|---|---|---|---|---|
| 1 |  |
-14.31 | 5 |  |
-0.25 |
| 2 |  |
-0.84 | 6 |  |
0.080 |
| 3 | 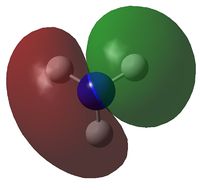 |
-0.45 | 7 | 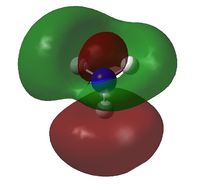 |
0.17 |
| 4 | 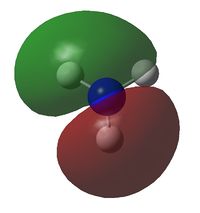 |
-0.45 | 8 | 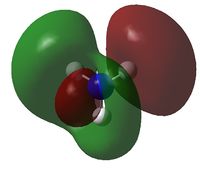 |
0.17 |
Charge Distribution
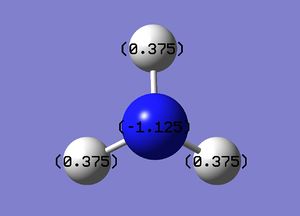
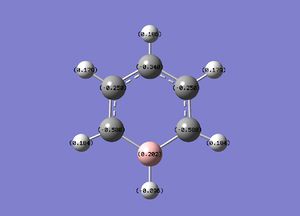
A natural bond orbital analysis was made of the .log file of ammonia. The charge distribution was calculated; red indicates a negatively chardged region and green a positively charged region. The colour range was from -1.000 to +1.000:
The relative charges were +0.375 for each of the three H atoms and -1.125 for the central N atom:
The natural charge numbers were found in the NBO analysis:
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
N 1 -1.12515 1.99982 6.11104 0.01429 8.12515
H 2 0.37505 0.00000 0.62250 0.00246 0.62495
H 3 0.37505 0.00000 0.62250 0.00246 0.62495
H 4 0.37505 0.00000 0.62249 0.00246 0.62495
=======================================================================
* Total * 0.00000 1.99982 7.97852 0.02166 10.00000
The NBO analysis separates the electron density into 2c2e bonds. It can be seen that for each N-H bond (Orbitals 1, 2, 3), 68.83% of the bond is contributed from orbitals on N; of these, the hybridisation character is 24.87% s orbital and 75.05% p orbital, indicating sp3. Orbital 4 is the core s orbital of the N atom with 100% s character. Orbital 5 is a lone pair which is formally occupied; its occupancy was calculated as 1.99721 - close to 2 electrons.
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.99909) BD ( 1) N 1 - H 2
( 68.83%) 0.8297* N 1 s( 24.87%)p 3.02( 75.05%)d 0.00( 0.09%)
-0.0001 -0.4986 -0.0059 0.0000 -0.2910
0.0052 0.8155 0.0277 0.0000 0.0000
0.0281 0.0000 0.0000 0.0032 0.0082
( 31.17%) 0.5583* H 2 s( 99.91%)p 0.00( 0.09%)
-0.9996 0.0000 0.0072 -0.0289 0.0000
2. (1.99909) BD ( 1) N 1 - H 3
( 68.83%) 0.8297* N 1 s( 24.86%)p 3.02( 75.05%)d 0.00( 0.09%)
0.0001 0.4986 0.0059 0.0000 0.2910
-0.0052 0.4077 0.0138 0.7062 0.0240
0.0140 0.0243 0.0076 0.0033 0.0031
( 31.17%) 0.5583* H 3 s( 99.91%)p 0.00( 0.09%)
0.9996 0.0000 -0.0072 -0.0145 -0.0250
3. (1.99909) BD ( 1) N 1 - H 4
( 68.83%) 0.8297* N 1 s( 24.87%)p 3.02( 75.05%)d 0.00( 0.09%)
0.0001 0.4986 0.0059 0.0000 0.2909
-0.0052 0.4077 0.0138 -0.7062 -0.0239
0.0140 -0.0243 -0.0076 0.0033 0.0031
( 31.17%) 0.5583* H 4 s( 99.91%)p 0.00( 0.09%)
0.9996 0.0000 -0.0072 -0.0145 0.0250
4. (1.99982) CR ( 1) N 1 s(100.00%)
1.0000 -0.0002 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
5. (1.99721) LP ( 1) N 1 s( 25.38%)p 2.94( 74.52%)d 0.00( 0.10%)
0.0001 0.5036 -0.0120 0.0000 -0.8618
0.0505 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 -0.0269 0.0155
The energy and population of the N-H bonds and N lone pair are shown below. Each of the N-H bonds are of identical occupancy and energy whilst the lone pair is relatively higher (less negative) in energy. The core s orbital of nitrogen is very low in energy relative to the valence orbitals.
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (H3N)
1. BD ( 1) N 1 - H 2 1.99909 -0.60417
2. BD ( 1) N 1 - H 3 1.99909 -0.60417
3. BD ( 1) N 1 - H 4 1.99909 -0.60416
4. CR ( 1) N 1 1.99982 -14.16768
5. LP ( 1) N 1 1.99721 -0.31756 24(v),16(v),20(v),17(v)
21(v),25(v)
NH3BH3
Optimisation
First the molecule was optimised: File:NH3BH3OPTCLU.LOG
Item Value Threshold Converged?
Maximum Force 0.000033 0.000450 YES
RMS Force 0.000019 0.000300 YES
Maximum Displacement 0.000439 0.001800 YES
RMS Displacement 0.000235 0.001200 YES
Predicted change in Energy=-2.479187D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,8) 1.0184 -DE/DX = 0.0 !
! R2 R(2,8) 1.0185 -DE/DX = 0.0 !
! R3 R(3,8) 1.0185 -DE/DX = 0.0 !
! R4 R(4,5) 1.2099 -DE/DX = 0.0 !
! R5 R(4,6) 1.2098 -DE/DX = 0.0 !
! R6 R(4,7) 1.2098 -DE/DX = 0.0 !
! R7 R(4,8) 1.6677 -DE/DX = 0.0 !
! A1 A(5,4,6) 113.8797 -DE/DX = 0.0 !
! A2 A(5,4,7) 113.8791 -DE/DX = 0.0 !
! A3 A(5,4,8) 104.5846 -DE/DX = 0.0 !
! A4 A(6,4,7) 113.8833 -DE/DX = 0.0 !
! A5 A(6,4,8) 104.5889 -DE/DX = 0.0 !
! A6 A(7,4,8) 104.5929 -DE/DX = 0.0 !
! A7 A(1,8,2) 107.885 -DE/DX = 0.0 !
! A8 A(1,8,3) 107.8862 -DE/DX = 0.0 !
! A9 A(1,8,4) 111.0218 -DE/DX = 0.0 !
! A10 A(2,8,3) 107.8918 -DE/DX = 0.0 !
! A11 A(2,8,4) 111.0077 -DE/DX = 0.0 !
! A12 A(3,8,4) 111.0059 -DE/DX = 0.0 !
! D1 D(5,4,8,1) 179.9831 -DE/DX = 0.0 !
! D2 D(5,4,8,2) -60.0164 -DE/DX = 0.0 !
! D3 D(5,4,8,3) 59.9822 -DE/DX = 0.0 !
! D4 D(6,4,8,1) -60.0199 -DE/DX = 0.0 !
! D5 D(6,4,8,2) 59.9806 -DE/DX = 0.0 !
! D6 D(6,4,8,3) 179.9793 -DE/DX = 0.0 !
! D7 D(7,4,8,1) 59.9849 -DE/DX = 0.0 !
! D8 D(7,4,8,2) 179.9855 -DE/DX = 0.0 !
! D9 D(7,4,8,3) -60.0159 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Frequency
A frequency analysis was then performed: File:NH3BH3FREQCLU.LOG
Item Value Threshold Converged?
Maximum Force 0.000004 0.000015 YES
RMS Force 0.000002 0.000010 YES
Maximum Displacement 0.000042 0.000060 YES
RMS Displacement 0.000018 0.000040 YES
Predicted change in Energy=-5.940207D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,8) 1.0185 -DE/DX = 0.0 !
! R2 R(2,8) 1.0185 -DE/DX = 0.0 !
! R3 R(3,8) 1.0185 -DE/DX = 0.0 !
! R4 R(4,5) 1.2098 -DE/DX = 0.0 !
! R5 R(4,6) 1.2098 -DE/DX = 0.0 !
! R6 R(4,7) 1.2098 -DE/DX = 0.0 !
! R7 R(4,8) 1.6677 -DE/DX = 0.0 !
! A1 A(5,4,6) 113.8743 -DE/DX = 0.0 !
! A2 A(5,4,7) 113.8742 -DE/DX = 0.0 !
! A3 A(5,4,8) 104.5968 -DE/DX = 0.0 !
! A4 A(6,4,7) 113.8745 -DE/DX = 0.0 !
! A5 A(6,4,8) 104.5968 -DE/DX = 0.0 !
! A6 A(7,4,8) 104.5968 -DE/DX = 0.0 !
! A7 A(1,8,2) 107.8733 -DE/DX = 0.0 !
! A8 A(1,8,3) 107.8733 -DE/DX = 0.0 !
! A9 A(1,8,4) 111.0254 -DE/DX = 0.0 !
! A10 A(2,8,3) 107.8734 -DE/DX = 0.0 !
! A11 A(2,8,4) 111.0254 -DE/DX = 0.0 !
! A12 A(3,8,4) 111.0253 -DE/DX = 0.0 !
! D1 D(5,4,8,1) 179.9993 -DE/DX = 0.0 !
! D2 D(5,4,8,2) -60.0007 -DE/DX = 0.0 !
! D3 D(5,4,8,3) 59.9994 -DE/DX = 0.0 !
! D4 D(6,4,8,1) -60.0007 -DE/DX = 0.0 !
! D5 D(6,4,8,2) 59.9992 -DE/DX = 0.0 !
! D6 D(6,4,8,3) 179.9993 -DE/DX = 0.0 !
! D7 D(7,4,8,1) 59.9995 -DE/DX = 0.0 !
! D8 D(7,4,8,2) 179.9994 -DE/DX = 0.0 !
! D9 D(7,4,8,3) -60.0005 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
The low frequencies were in a narrow range; no negative values were reported.
Low frequencies --- -2.8627 0.0003 0.0005 0.0006 1.2904 4.1827 Low frequencies --- 263.4385 632.9764 638.4467
| Mode | Frequency (cm-1) | Infrared | Description | Symmetry Point Group (C1) |
|---|---|---|---|---|
| 1 | 263 | 0 | Wagging about B-N bond | A |
| 2 | 633 | 14 | B-N stretching | A |
| 3 | 638 | 4 | B-N bending | A |
| 4 | 638 | 34 | B-N bending | A |
| 5 | 1069 | 41 | B-N see-sawing | A |
| 6 | 1069 | 41 | B-N see-sawing | A |
| 7 | 1196 | 109 | 3H of BH3 concerted wagging in and out of plane; cf umbrella opening/closing | A |
| 8 | 1204 | 3 | 3H of BH3 asymmetric bending | A |
| 9 | 1204 | 3 | 3H of BH3 asymmetric bending | A |
| 10 | 1329 | 114 | 3H of NH3 concerted wagging in and out of plane; cf umbrella opening/closing | A |
| 11 | 1676 | 28 | 3H of NH3 asymmetric bending | A |
| 12 | 1676 | 28 | 3H of NH3 asymmetric bending | A |
| 13 | 2472 | 67 | 3H of BH3 concerted symmetric stretching | A |
| 14 | 2532 | 231 | 3H of BH3 asymmetric stretching | A |
| 15 | 2532 | 231 | 3H of BH3 asymmetric stretching | A |
| 16 | 3464 | 3 | 3H of NH3 concerted symmetric stretching | A |
| 17 | 3581 | 28 | 3H of NH3 asymmetric stretching | A |
| 18 | 3581 | 28 | 3H of NH3 asymmetric stretching | A |
Mode 1 is the only infrared inactive mode.
Association Energy of Ammonia-Borane
- E(BH3) = -26.61532364 a.u.
- E(NH3) = -56.55776872 a.u.
- E(BH3NH3) = -83.22468909 a.u.
ΔE = E(NH3BH3)-[E(NH3)+E(BH3)]
ΔE = -83.22468909 a.u. - -83.17309236
ΔE = -0.05159673 a.u.
1 a.u. = 2625.50 kJ mol-1
Enthalpy of formation of ammonia-borane ΔHf = -0.05159673 a.u. * 2625.50 = -135.467214615 kJ mol-1 = -135.5 kJ mol-1
This calculated value is somewhat lower than that found experimentally of -172.1 kJ mol-1[3].
Aromaticity Mini-Project
The molecular orbitals, LCAOs and energies of benzene and its isoelectronic analogues boratabenzene, pyridinium ion and borazine were investigated. All calculations were performed on the HPC and files published in D-space.
Benzene
A molecule of benzene was optimised with a 63G1(d,p) basis set: [5]
Item Value Threshold Converged?
Maximum Force 0.000212 0.000450 YES
RMS Force 0.000085 0.000300 YES
Maximum Displacement 0.000991 0.001800 YES
RMS Displacement 0.000315 0.001200 YES
Predicted change in Energy=-5.157454D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3963 -DE/DX = 0.0001 !
! R2 R(1,6) 1.3961 -DE/DX = 0.0002 !
! R3 R(1,7) 1.0861 -DE/DX = 0.0002 !
! R4 R(2,3) 1.3961 -DE/DX = 0.0002 !
! R5 R(2,8) 1.0861 -DE/DX = 0.0002 !
! R6 R(3,4) 1.3963 -DE/DX = 0.0001 !
! R7 R(3,9) 1.086 -DE/DX = 0.0002 !
! R8 R(4,5) 1.3961 -DE/DX = 0.0002 !
! R9 R(4,10) 1.086 -DE/DX = 0.0002 !
! R10 R(5,6) 1.3963 -DE/DX = 0.0001 !
! R11 R(5,11) 1.0861 -DE/DX = 0.0002 !
! R12 R(6,12) 1.0861 -DE/DX = 0.0002 !
! A1 A(2,1,6) 119.9972 -DE/DX = 0.0 !
! A2 A(2,1,7) 119.9949 -DE/DX = 0.0 !
! A3 A(6,1,7) 120.0079 -DE/DX = 0.0 !
! A4 A(1,2,3) 120.0079 -DE/DX = 0.0 !
! A5 A(1,2,8) 119.9881 -DE/DX = 0.0 !
! A6 A(3,2,8) 120.004 -DE/DX = 0.0 !
! A7 A(2,3,4) 119.9948 -DE/DX = 0.0 !
! A8 A(2,3,9) 120.0086 -DE/DX = 0.0 !
! A9 A(4,3,9) 119.9966 -DE/DX = 0.0 !
! A10 A(3,4,5) 119.9972 -DE/DX = 0.0 !
! A11 A(3,4,10) 119.9934 -DE/DX = 0.0 !
! A12 A(5,4,10) 120.0094 -DE/DX = 0.0 !
! A13 A(4,5,6) 120.0083 -DE/DX = 0.0 !
! A14 A(4,5,11) 120.0014 -DE/DX = 0.0 !
! A15 A(6,5,11) 119.9904 -DE/DX = 0.0 !
! A16 A(1,6,5) 119.9946 -DE/DX = 0.0 !
! A17 A(1,6,12) 120.0106 -DE/DX = 0.0 !
! A18 A(5,6,12) 119.9948 -DE/DX = 0.0 !
! D1 D(6,1,2,3) -0.0059 -DE/DX = 0.0 !
! D2 D(6,1,2,8) 180.0023 -DE/DX = 0.0 !
! D3 D(7,1,2,3) -180.01 -DE/DX = 0.0 !
! D4 D(7,1,2,8) -0.0019 -DE/DX = 0.0 !
! D5 D(2,1,6,5) -0.0055 -DE/DX = 0.0 !
! D6 D(2,1,6,12) -179.9972 -DE/DX = 0.0 !
! D7 D(7,1,6,5) -180.0013 -DE/DX = 0.0 !
! D8 D(7,1,6,12) 0.007 -DE/DX = 0.0 !
! D9 D(1,2,3,4) 0.0117 -DE/DX = 0.0 !
! D10 D(1,2,3,9) -179.9914 -DE/DX = 0.0 !
! D11 D(8,2,3,4) 180.0036 -DE/DX = 0.0 !
! D12 D(8,2,3,9) 0.0005 -DE/DX = 0.0 !
! D13 D(2,3,4,5) -0.0062 -DE/DX = 0.0 !
! D14 D(2,3,4,10) -180.0059 -DE/DX = 0.0 !
! D15 D(9,3,4,5) 179.9969 -DE/DX = 0.0 !
! D16 D(9,3,4,10) -0.0028 -DE/DX = 0.0 !
! D17 D(3,4,5,6) -0.0051 -DE/DX = 0.0 !
! D18 D(3,4,5,11) 180.0058 -DE/DX = 0.0 !
! D19 D(10,4,5,6) -180.0055 -DE/DX = 0.0 !
! D20 D(10,4,5,11) 0.0054 -DE/DX = 0.0 !
! D21 D(4,5,6,1) 0.011 -DE/DX = 0.0 !
! D22 D(4,5,6,12) 180.0027 -DE/DX = 0.0 !
! D23 D(11,5,6,1) -179.9999 -DE/DX = 0.0 !
! D24 D(11,5,6,12) -0.0082 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Frequency
A frequency analysis was then performed: [6]
Item Value Threshold Converged?
Maximum Force 0.000005 0.000015 YES
RMS Force 0.000002 0.000010 YES
Maximum Displacement 0.000035 0.000060 YES
RMS Displacement 0.000014 0.000040 YES
Predicted change in Energy=-5.496302D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3963 -DE/DX = 0.0 !
! R2 R(1,6) 1.3963 -DE/DX = 0.0 !
! R3 R(1,7) 1.0863 -DE/DX = 0.0 !
! R4 R(2,3) 1.3963 -DE/DX = 0.0 !
! R5 R(2,8) 1.0863 -DE/DX = 0.0 !
! R6 R(3,4) 1.3963 -DE/DX = 0.0 !
! R7 R(3,9) 1.0863 -DE/DX = 0.0 !
! R8 R(4,5) 1.3963 -DE/DX = 0.0 !
! R9 R(4,10) 1.0863 -DE/DX = 0.0 !
! R10 R(5,6) 1.3963 -DE/DX = 0.0 !
! R11 R(5,11) 1.0863 -DE/DX = 0.0 !
! R12 R(6,12) 1.0863 -DE/DX = 0.0 !
! A1 A(2,1,6) 120.0004 -DE/DX = 0.0 !
! A2 A(2,1,7) 120.0 -DE/DX = 0.0 !
! A3 A(6,1,7) 119.9995 -DE/DX = 0.0 !
! A4 A(1,2,3) 119.9991 -DE/DX = 0.0 !
! A5 A(1,2,8) 120.0004 -DE/DX = 0.0 !
! A6 A(3,2,8) 120.0005 -DE/DX = 0.0 !
! A7 A(2,3,4) 120.0005 -DE/DX = 0.0 !
! A8 A(2,3,9) 120.0 -DE/DX = 0.0 !
! A9 A(4,3,9) 119.9995 -DE/DX = 0.0 !
! A10 A(3,4,5) 120.0004 -DE/DX = 0.0 !
! A11 A(3,4,10) 119.9993 -DE/DX = 0.0 !
! A12 A(5,4,10) 120.0003 -DE/DX = 0.0 !
! A13 A(4,5,6) 119.9991 -DE/DX = 0.0 !
! A14 A(4,5,11) 120.0007 -DE/DX = 0.0 !
! A15 A(6,5,11) 120.0003 -DE/DX = 0.0 !
! A16 A(1,6,5) 120.0005 -DE/DX = 0.0 !
! A17 A(1,6,12) 119.9998 -DE/DX = 0.0 !
! A18 A(5,6,12) 119.9997 -DE/DX = 0.0 !
! D1 D(6,1,2,3) 0.0001 -DE/DX = 0.0 !
! D2 D(6,1,2,8) 180.0 -DE/DX = 0.0 !
! D3 D(7,1,2,3) 180.0002 -DE/DX = 0.0 !
! D4 D(7,1,2,8) 0.0001 -DE/DX = 0.0 !
! D5 D(2,1,6,5) 0.0001 -DE/DX = 0.0 !
! D6 D(2,1,6,12) 179.9999 -DE/DX = 0.0 !
! D7 D(7,1,6,5) 180.0001 -DE/DX = 0.0 !
! D8 D(7,1,6,12) -0.0002 -DE/DX = 0.0 !
! D9 D(1,2,3,4) -0.0003 -DE/DX = 0.0 !
! D10 D(1,2,3,9) 179.9999 -DE/DX = 0.0 !
! D11 D(8,2,3,4) 179.9998 -DE/DX = 0.0 !
! D12 D(8,2,3,9) 0.0 -DE/DX = 0.0 !
! D13 D(2,3,4,5) 0.0001 -DE/DX = 0.0 !
! D14 D(2,3,4,10) -179.9998 -DE/DX = 0.0 !
! D15 D(9,3,4,5) 180.0 -DE/DX = 0.0 !
! D16 D(9,3,4,10) 0.0001 -DE/DX = 0.0 !
! D17 D(3,4,5,6) 0.0001 -DE/DX = 0.0 !
! D18 D(3,4,5,11) -180.0001 -DE/DX = 0.0 !
! D19 D(10,4,5,6) 180.0001 -DE/DX = 0.0 !
! D20 D(10,4,5,11) -0.0002 -DE/DX = 0.0 !
! D21 D(4,5,6,1) -0.0002 -DE/DX = 0.0 !
! D22 D(4,5,6,12) 180.0 -DE/DX = 0.0 !
! D23 D(11,5,6,1) 180.0 -DE/DX = 0.0 !
! D24 D(11,5,6,12) 0.0002 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
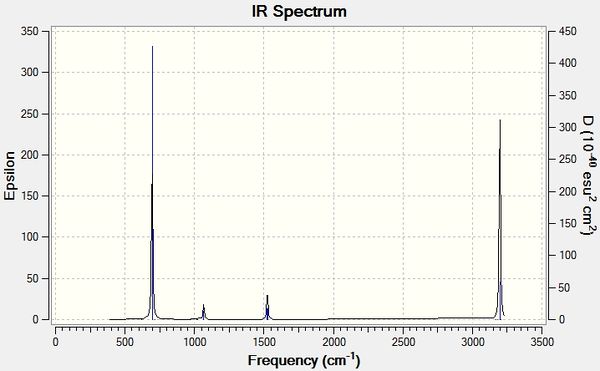
Low frequencies --- -0.0009 -0.0006 0.0008 1.2764 2.8520 9.2722 Low frequencies --- 414.5347 414.6189 621.0767
The calculation converged successfully; no negative values for the low frequencies.
| Mode | Frequency (cm-1) | Infrared | Description | Symmetry Point Group (C1) |
|---|---|---|---|---|
| 1 | 414 | 0 | 2 pairs of opposite Hs wagging alternately in and out of plane | A |
| 2 | 414 | 0 | 2 pairs of opposite Hs wagging alternately in and out of plane | A |
| 3 | 638 | 4 | C-C in-plane bend | A |
| 4 | 638 | 4 | C-C in-plane bend | A |
| 5 | 695 | 74 | C-H in and out of plane wagging | A |
| 6 | 718 | 41 | Alternating C-C in and out of plane see-sawing | A |
| 7 | 865 | 0 | 2 pairs of opposite Hs concerted wagging in and out of plane | A |
| 8 | 865 | 0 | 2 pairs of opposite Hs concerted wagging in and out of plane | A |
| 9 | 975 | 0 | 2 pairs of opposite Hs alternate wagging in and out of plane | A |
| 10 | 975 | 0 | 2 pairs of opposite Hs alternate wagging in and out of plane | A |
| 11 | 1013 | 0 | Alternate C-H in and out of plane wagging | A |
| 12 | 1018 | 0 | C-C in-plane alternating asymmetric stretching | A |
| 13 | 1020 | 0 | C-C in-plane symmetric stretching | A |
| 14 | 1066 | 3 | C-C in-plane asymmetric stretching | A |
| 15 | 1067 | 3 | C-C in-plane asymmetric stretching | A |
| 16 | 1180 | 0 | C-H in-plane asymmetric bending | A |
| 17 | 1202 | 0 | 2 pairs of opposite Hs C-H in-plane asymmetric bending | A |
| 18 | 1203 | 0 | C-H in-plane asymmetric bending | A |
| 19 | 1356 | 0 | C-C in-plane asymmetric stretching | A |
| 20 | 1381 | 0 | C-H in-plane symmetric bending | A |
| 21 | 1524 | 7 | C-C, C-H in-plane alternating stretching | A |
| 22 | 1525 | 7 | C-C, C-H in-plane alternating stretching | A |
| 23 | 1653 | 0 | C-C in-plane asymmetric stretching | A |
| 24 | 1653 | 0 | C-C in-plane asymmetric stretching | A |
| 25 | 3172 | 0 | Alternating C-H in-plane stretching | A |
| 26 | 3181 | 0 | 2 pairs of opposite Hs in-plane asymmetric stretching | A |
| 27 | 3181 | 0 | 2 pairs of opposite Hs alternating to 1 pair in-plane stretching | A |
| 28 | 3197 | 47 | 2 pairs of opposite Hs asymmetric stretching | A |
| 29 | 3197 | 47 | 1 half of 3H asymmetric in-plane stretching | A |
| 30 | 3208 | 0 | 6H symmetric in-plane stretching | A |
| 13 | 1020 | 0 | C-C in-plane symmetric stretching | A |
| 14 | 1066 | 3 | C-C in-plane asymmetric stretching | A |
| 15 | 1067 | 3 | C-C in-plane asymmetric stretching | A |
Only modes 5, 6 (degenerate pair), 14, 15 (degenerate pair), 21, 22 (degenerate pair)) and 28, 29 (degenerate pair) are intense enough to be observed on the spectrum. The remainder either do not result ina change in dipole moment, or the dipole moment is too weak to be of sufficient intensity to be visible on the spectrum.
NBO Analysis
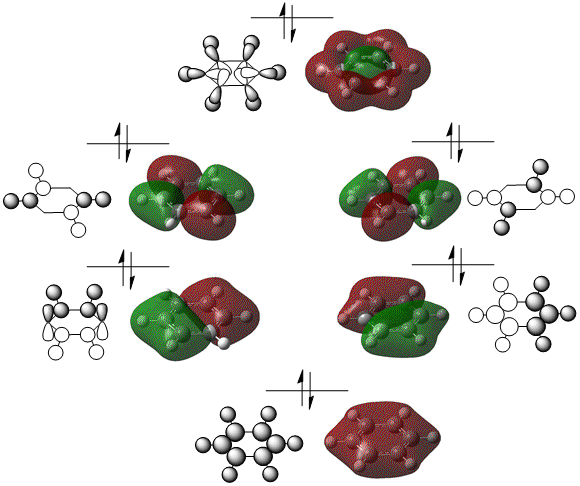
The MOs were computed [7]. The first 6 MOs by energy are core molecular orbitals and involve the 2s orbitals of the carbon atoms which do not affect the stability of the whole molecule to a significant degree. Below are shown the 7th MO up to the doubly degenerate LUMO.
| MO | Image | MO | Image | MO | Image | MO | Image | MO | Image | MO | Image |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 7 |  |
10 | 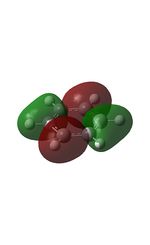 |
13 |  |
16 |  |
19 | 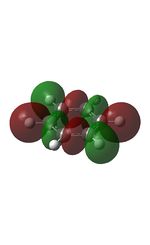 |
22 | 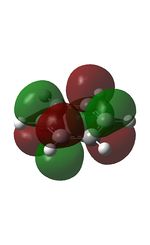
|
| 8 |  |
11 |  |
14 |  |
17 |  |
20 | 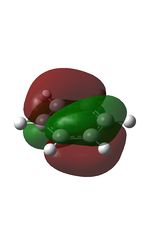 |
23 | 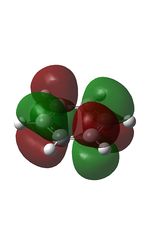
|
| 9 |  |
12 |  |
15 |  |
18 | 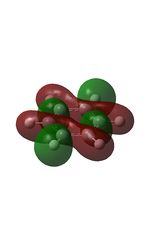 |
21 | 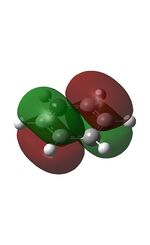
|
The MOs of benzene, both sigma and pi, were used to infer the LCAOs.
It can be seen from the NBO analysis that the molecule is symmetrical and all C-H bonds are identical; all C atoms have the same electron density, as do all H atoms. The MOs reinforce the concept of aromaticity as it is visible from the totally pi bonding MO that the electron density covers the entire surface equally above and below the ring plane, indicating delocalisation of the pi electrons; the probability of finding an electron anywhere in pi system is equal. Were the pi electrons localised, the overlap of the ring of p orbitals would not extend to the centre of the ring and there would be a region of 0 electron density at the centre.
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
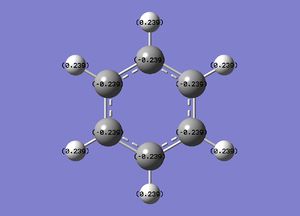
C 1 -0.23858 1.99910 4.22617 0.01331 6.23858
C 2 -0.23858 1.99910 4.22617 0.01331 6.23858
C 3 -0.23862 1.99910 4.22620 0.01331 6.23862
C 4 -0.23858 1.99910 4.22617 0.01331 6.23858
C 5 -0.23858 1.99910 4.22617 0.01331 6.23858
C 6 -0.23862 1.99910 4.22620 0.01331 6.23862
H 7 0.23859 0.00000 0.75997 0.00144 0.76141
H 8 0.23859 0.00000 0.75997 0.00144 0.76141
H 9 0.23860 0.00000 0.75997 0.00144 0.76140
H 10 0.23859 0.00000 0.75997 0.00144 0.76141
H 11 0.23859 0.00000 0.75997 0.00144 0.76141
H 12 0.23860 0.00000 0.75997 0.00144 0.76140
=======================================================================
* Total * 0.00000 11.99462 29.91692 0.08846 42.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98098) BD ( 1) C 1 - C 2
( 50.00%) 0.7071* C 1 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
( 50.00%) 0.7071* C 2 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
2. (1.98098) BD ( 1) C 1 - C 6
( 50.00%) 0.7071* C 1 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
( 50.00%) 0.7071* C 6 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
3. (1.66515) BD ( 2) C 1 - C 6
( 50.00%) 0.7071* C 1 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
( 50.00%) 0.7071* C 6 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
4. (1.98306) BD ( 1) C 1 - H 7
( 62.04%) 0.7877* C 1 s( 29.57%)p 2.38( 70.39%)d 0.00( 0.04%)
( 37.96%) 0.6161* H 7 s( 99.95%)p 0.00( 0.05%)
5. (1.98098) BD ( 1) C 2 - C 3
( 50.00%) 0.7071* C 2 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
( 50.00%) 0.7071* C 3 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
6. (1.66515) BD ( 2) C 2 - C 3
( 50.00%) 0.7071* C 2 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
( 50.00%) 0.7071* C 3 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
7. (1.98306) BD ( 1) C 2 - H 8
( 62.04%) 0.7877* C 2 s( 29.57%)p 2.38( 70.39%)d 0.00( 0.04%)
( 37.96%) 0.6161* H 8 s( 99.95%)p 0.00( 0.05%)
8. (1.98098) BD ( 1) C 3 - C 4
( 50.00%) 0.7071* C 3 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
( 50.00%) 0.7071* C 4 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
9. (1.98306) BD ( 1) C 3 - H 9
( 62.04%) 0.7877* C 3 s( 29.57%)p 2.38( 70.39%)d 0.00( 0.04%)
( 37.96%) 0.6161* H 9 s( 99.95%)p 0.00( 0.05%)
10. (1.98098) BD ( 1) C 4 - C 5
( 50.00%) 0.7071* C 4 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
( 50.00%) 0.7071* C 5 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
11. (1.66513) BD ( 2) C 4 - C 5
( 50.00%) 0.7071* C 4 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
( 50.00%) 0.7071* C 5 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
12. (1.98306) BD ( 1) C 4 - H 10
( 62.04%) 0.7877* C 4 s( 29.57%)p 2.38( 70.39%)d 0.00( 0.04%)
( 37.96%) 0.6161* H 10 s( 99.95%)p 0.00( 0.05%)
13. (1.98098) BD ( 1) C 5 - C 6
( 50.00%) 0.7071* C 5 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
( 50.00%) 0.7071* C 6 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
14. (1.98306) BD ( 1) C 5 - H 11
( 62.04%) 0.7877* C 5 s( 29.57%)p 2.38( 70.39%)d 0.00( 0.04%)
( 37.96%) 0.6161* H 11 s( 99.95%)p 0.00( 0.05%)
15. (1.98306) BD ( 1) C 6 - H 12
( 62.04%) 0.7877* C 6 s( 29.57%)p 2.38( 70.39%)d 0.00( 0.04%)
( 37.96%) 0.6161* H 12 s( 99.95%)p 0.00( 0.05%)
16. (1.99911) CR ( 1) C 1 s(100.00%)p 0.00( 0.00%)
17. (1.99911) CR ( 1) C 2 s(100.00%)p 0.00( 0.00%)
18. (1.99911) CR ( 1) C 3 s(100.00%)p 0.00( 0.00%)
19. (1.99911) CR ( 1) C 4 s(100.00%)p 0.00( 0.00%)
20. (1.99911) CR ( 1) C 5 s(100.00%)p 0.00( 0.00%)
21. (1.99911) CR ( 1) C 6 s(100.00%)p 0.00( 0.00%)
It can be seen that all C atoms have a hybrid sp2 orbital and a single p orbital perpendicular to the ring plane, reinforcing the concept of aromaticity.
Natural Bond Orbitals (Summary):
NBO Occupancy Energy
===========================================================
Molecular unit 1 (C6H6)
1. BD ( 1) C 1 - C 2 1.98098 -0.68189
2. BD ( 1) C 1 - C 6 1.98098 -0.68190
3. BD ( 2) C 1 - C 6 1.66515 -0.23791
5. BD ( 1) C 2 - C 3 1.98098 -0.68190
6. BD ( 2) C 2 - C 3 1.66515 -0.23791
8. BD ( 1) C 3 - C 4 1.98098 -0.68190
9. BD ( 1) C 3 - H 9 1.98306 -0.51228
10. BD ( 1) C 4 - C 5 1.98098 -0.68189
11. BD ( 2) C 4 - C 5 1.66513 -0.23791
13. BD ( 1) C 5 - C 6 1.98098 -0.68190
14. BD ( 1) C 5 - H 11 1.98306 -0.51228
15. BD ( 1) C 6 - H 12 1.98306 -0.51228
16. CR ( 1) C 1 1.99911 -10.04069
17. CR ( 1) C 2 1.99911 -10.04069
18. CR ( 1) C 3 1.99911 -10.04069
19. CR ( 1) C 4 1.99911 -10.04069
20. CR ( 1) C 5 1.99911 -10.04069
21. CR ( 1) C 6 1.99911 -10.04069
The charge distribution ranges from -1.000 to +1.000:
Again it can be seen that all H atoms have the same electron distribution, as do all C atoms.
Boratabenzene
Boratabenzene is a benzene analogue with one C-H unit replaced by B-H. To be isoelectronic with benzene, a -1 charge is applied to the molecule.
Optimisation
Published in D-space: [8]
Item Value Threshold Converged?
Maximum Force 0.000159 0.000450 YES
RMS Force 0.000069 0.000300 YES
Maximum Displacement 0.000911 0.001800 YES
RMS Displacement 0.000335 0.001200 YES
Predicted change in Energy=-6.630178D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3989 -DE/DX = 0.0 !
! R2 R(1,6) 1.097 -DE/DX = -0.0001 !
! R3 R(1,12) 1.5137 -DE/DX = 0.0001 !
! R4 R(2,3) 1.4053 -DE/DX = -0.0001 !
! R5 R(2,7) 1.0968 -DE/DX = 0.0001 !
! R6 R(3,4) 1.4053 -DE/DX = -0.0001 !
! R7 R(3,8) 1.0917 -DE/DX = -0.0001 !
! R8 R(4,5) 1.3989 -DE/DX = 0.0 !
! R9 R(4,9) 1.0968 -DE/DX = 0.0001 !
! R10 R(5,10) 1.097 -DE/DX = -0.0001 !
! R11 R(5,12) 1.5138 -DE/DX = 0.0001 !
! R12 R(11,12) 1.2185 -DE/DX = 0.0 !
! A1 A(2,1,6) 115.9494 -DE/DX = 0.0001 !
! A2 A(2,1,12) 120.0809 -DE/DX = -0.0001 !
! A3 A(6,1,12) 123.9697 -DE/DX = -0.0001 !
! A4 A(1,2,3) 122.1393 -DE/DX = 0.0001 !
! A5 A(1,2,7) 120.4237 -DE/DX = -0.0002 !
! A6 A(3,2,7) 117.437 -DE/DX = 0.0 !
! A7 A(2,3,4) 120.4508 -DE/DX = -0.0001 !
! A8 A(2,3,8) 119.776 -DE/DX = 0.0001 !
! A9 A(4,3,8) 119.7732 -DE/DX = 0.0001 !
! A10 A(3,4,5) 122.1381 -DE/DX = 0.0001 !
! A11 A(3,4,9) 117.4352 -DE/DX = 0.0 !
! A12 A(5,4,9) 120.4266 -DE/DX = -0.0002 !
! A13 A(4,5,10) 115.9535 -DE/DX = 0.0001 !
! A14 A(4,5,12) 120.0812 -DE/DX = -0.0001 !
! A15 A(10,5,12) 123.9653 -DE/DX = -0.0001 !
! A16 A(1,12,5) 115.1096 -DE/DX = 0.0 !
! A17 A(1,12,11) 122.4482 -DE/DX = 0.0 !
! A18 A(5,12,11) 122.4422 -DE/DX = 0.0 !
! D1 D(6,1,2,3) 180.01 -DE/DX = 0.0 !
! D2 D(6,1,2,7) -0.0042 -DE/DX = 0.0 !
! D3 D(12,1,2,3) 0.0084 -DE/DX = 0.0 !
! D4 D(12,1,2,7) -180.0058 -DE/DX = 0.0 !
! D5 D(2,1,12,5) 0.001 -DE/DX = 0.0 !
! D6 D(2,1,12,11) -180.0 -DE/DX = 0.0 !
! D7 D(6,1,12,5) -180.0007 -DE/DX = 0.0 !
! D8 D(6,1,12,11) -0.0017 -DE/DX = 0.0 !
! D9 D(1,2,3,4) -0.015 -DE/DX = 0.0 !
! D10 D(1,2,3,8) -180.0108 -DE/DX = 0.0 !
! D11 D(7,2,3,4) -180.0011 -DE/DX = 0.0 !
! D12 D(7,2,3,8) 0.0031 -DE/DX = 0.0 !
! D13 D(2,3,4,5) 0.0116 -DE/DX = 0.0 !
! D14 D(2,3,4,9) 180.0045 -DE/DX = 0.0 !
! D15 D(8,3,4,5) 180.0075 -DE/DX = 0.0 !
! D16 D(8,3,4,9) 0.0004 -DE/DX = 0.0 !
! D17 D(3,4,5,10) -180.0055 -DE/DX = 0.0 !
! D18 D(3,4,5,12) -0.002 -DE/DX = 0.0 !
! D19 D(9,4,5,10) 0.0018 -DE/DX = 0.0 !
! D20 D(9,4,5,12) 180.0053 -DE/DX = 0.0 !
! D21 D(4,5,12,1) -0.0042 -DE/DX = 0.0 !
! D22 D(4,5,12,11) -180.0032 -DE/DX = 0.0 !
! D23 D(10,5,12,1) 179.9996 -DE/DX = 0.0 !
! D24 D(10,5,12,11) 0.0006 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Frequency
Published in D-space: [9]
Item Value Threshold Converged?
Maximum Force 0.000002 0.000015 YES
RMS Force 0.000001 0.000010 YES
Maximum Displacement 0.000035 0.000060 YES
RMS Displacement 0.000010 0.000040 YES
Predicted change in Energy=-1.395415D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3988 -DE/DX = 0.0 !
! R2 R(1,6) 1.0968 -DE/DX = 0.0 !
! R3 R(1,12) 1.5141 -DE/DX = 0.0 !
! R4 R(2,3) 1.4051 -DE/DX = 0.0 !
! R5 R(2,7) 1.0969 -DE/DX = 0.0 !
! R6 R(3,4) 1.4052 -DE/DX = 0.0 !
! R7 R(3,8) 1.0915 -DE/DX = 0.0 !
! R8 R(4,5) 1.3988 -DE/DX = 0.0 !
! R9 R(4,9) 1.0969 -DE/DX = 0.0 !
! R10 R(5,10) 1.0968 -DE/DX = 0.0 !
! R11 R(5,12) 1.5141 -DE/DX = 0.0 !
! R12 R(11,12) 1.2184 -DE/DX = 0.0 !
! A1 A(2,1,6) 116.0114 -DE/DX = 0.0 !
! A2 A(2,1,12) 120.0605 -DE/DX = 0.0 !
! A3 A(6,1,12) 123.9281 -DE/DX = 0.0 !
! A4 A(1,2,3) 122.18 -DE/DX = 0.0 !
! A5 A(1,2,7) 120.3636 -DE/DX = 0.0 !
! A6 A(3,2,7) 117.4564 -DE/DX = 0.0 !
! A7 A(2,3,4) 120.415 -DE/DX = 0.0 !
! A8 A(2,3,8) 119.7926 -DE/DX = 0.0 !
! A9 A(4,3,8) 119.7924 -DE/DX = 0.0 !
! A10 A(3,4,5) 122.1798 -DE/DX = 0.0 !
! A11 A(3,4,9) 117.4563 -DE/DX = 0.0 !
! A12 A(5,4,9) 120.364 -DE/DX = 0.0 !
! A13 A(4,5,10) 116.0119 -DE/DX = 0.0 !
! A14 A(4,5,12) 120.0607 -DE/DX = 0.0 !
! A15 A(10,5,12) 123.9275 -DE/DX = 0.0 !
! A16 A(1,12,5) 115.1041 -DE/DX = 0.0 !
! A17 A(1,12,11) 122.4484 -DE/DX = 0.0 !
! A18 A(5,12,11) 122.4475 -DE/DX = 0.0 !
! D1 D(6,1,2,3) 179.9996 -DE/DX = 0.0 !
! D2 D(6,1,2,7) 0.0001 -DE/DX = 0.0 !
! D3 D(12,1,2,3) 0.0005 -DE/DX = 0.0 !
! D4 D(12,1,2,7) -179.999 -DE/DX = 0.0 !
! D5 D(2,1,12,5) -0.002 -DE/DX = 0.0 !
! D6 D(2,1,12,11) 180.0006 -DE/DX = 0.0 !
! D7 D(6,1,12,5) -180.001 -DE/DX = 0.0 !
! D8 D(6,1,12,11) 0.0016 -DE/DX = 0.0 !
! D9 D(1,2,3,4) 0.0011 -DE/DX = 0.0 !
! D10 D(1,2,3,8) -179.9996 -DE/DX = 0.0 !
! D11 D(7,2,3,4) -179.9994 -DE/DX = 0.0 !
! D12 D(7,2,3,8) -0.0001 -DE/DX = 0.0 !
! D13 D(2,3,4,5) -0.0011 -DE/DX = 0.0 !
! D14 D(2,3,4,9) 179.9993 -DE/DX = 0.0 !
! D15 D(8,3,4,5) 179.9996 -DE/DX = 0.0 !
! D16 D(8,3,4,9) 0.0 -DE/DX = 0.0 !
! D17 D(3,4,5,10) -179.9997 -DE/DX = 0.0 !
! D18 D(3,4,5,12) -0.0006 -DE/DX = 0.0 !
! D19 D(9,4,5,10) 0.0 -DE/DX = 0.0 !
! D20 D(9,4,5,12) 179.999 -DE/DX = 0.0 !
! D21 D(4,5,12,1) 0.0021 -DE/DX = 0.0 !
! D22 D(4,5,12,11) 179.9995 -DE/DX = 0.0 !
! D23 D(10,5,12,1) 180.0011 -DE/DX = 0.0 !
! D24 D(10,5,12,11) -0.0015 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
All low frequencies were in a narrow range and there were no positive values.
Low frequencies --- -7.1559 -0.0008 -0.0005 -0.0003 3.4577 4.6551 Low frequencies --- 371.2990 404.4178 565.0787
MO Analysis
The MOs were computed and published in D-space: [10]
| MO | Image | MO | Image | MO | Image | MO | Image | MO | Image | MO | Image |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 7 | 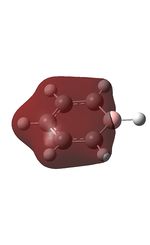 |
10 | 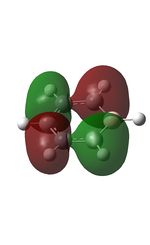 |
13 | 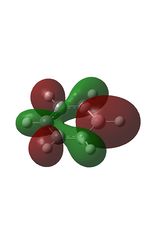 |
16 |  |
19 |  |
22 | 
|
| 8 |  |
11 |  |
14 |  |
17 |  |
20 |  |
23 | 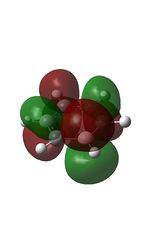
|
| 9 | 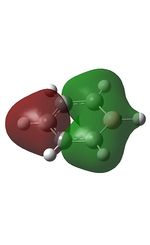 |
12 | 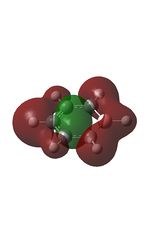 |
15 | 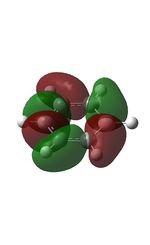 |
18 | 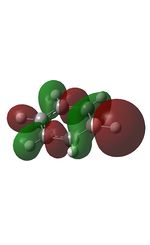 |
21 | 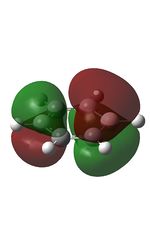
|
NBO analysis
NBO analysis showed that unlike benzene, electron distribution was no longer equal amongst all C atoms:
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
C 1 -0.58804 1.99901 4.57724 0.01178 6.58804
C 2 -0.25033 1.99910 4.23710 0.01413 6.25033
C 3 -0.34002 1.99907 4.32711 0.01384 6.34002
C 4 -0.25033 1.99910 4.23710 0.01413 6.25033
C 5 -0.58803 1.99901 4.57724 0.01178 6.58803
H 6 0.18385 0.00000 0.81397 0.00218 0.81615
H 7 0.17899 0.00000 0.81839 0.00263 0.82101
H 8 0.18574 0.00000 0.81227 0.00199 0.81426
H 9 0.17899 0.00000 0.81839 0.00263 0.82101
H 10 0.18385 0.00000 0.81397 0.00218 0.81615
H 11 -0.09648 0.00000 1.09595 0.00054 1.09648
B 12 0.20182 1.99906 2.78752 0.01160 4.79818
=======================================================================
* Total * -1.00000 11.99436 29.91623 0.08941 42.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98271) BD ( 1) C 1 - C 2
( 49.23%) 0.7017* C 1 s( 32.50%)p 2.08( 67.45%)d 0.00( 0.05%)
( 50.77%) 0.7125* C 2 s( 37.60%)p 1.66( 62.37%)d 0.00( 0.03%)
2. (1.76873) BD ( 2) C 1 - C 2
( 51.88%) 0.7202* C 1 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
( 48.12%) 0.6937* C 2 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
3. (1.98422) BD ( 1) C 1 - H 6
( 59.41%) 0.7708* C 1 s( 25.40%)p 2.93( 74.55%)d 0.00( 0.05%)
( 40.59%) 0.6371* H 6 s( 99.95%)p 0.00( 0.05%)
-0.9998 -0.0005 0.0191 -0.0100 0.0000
4. (1.96998) BD ( 1) C 1 - B 12
( 66.70%) 0.8167* C 1 s( 42.00%)p 1.38( 57.99%)d 0.00( 0.01%)
( 33.30%) 0.5771* B 12 s( 33.40%)p 1.99( 66.53%)d 0.00( 0.08%)
5. (1.97969) BD ( 1) C 2 - C 3
( 49.96%) 0.7068* C 2 s( 35.53%)p 1.81( 64.43%)d 0.00( 0.04%)
( 50.04%) 0.7074* C 3 s( 35.87%)p 1.79( 64.10%)d 0.00( 0.04%)
6. (1.98568) BD ( 1) C 2 - H 7
( 59.32%) 0.7702* C 2 s( 26.85%)p 2.72( 73.10%)d 0.00( 0.05%)
( 40.68%) 0.6378* H 7 s( 99.95%)p 0.00( 0.05%)
7. (1.97969) BD ( 1) C 3 - C 4
( 50.04%) 0.7074* C 3 s( 35.87%)p 1.79( 64.10%)d 0.00( 0.04%)
( 49.96%) 0.7068* C 4 s( 35.53%)p 1.81( 64.43%)d 0.00( 0.04%)
8. (1.98505) BD ( 1) C 3 - H 8
( 59.44%) 0.7710* C 3 s( 28.24%)p 2.54( 71.72%)d 0.00( 0.04%)
( 40.56%) 0.6369* H 8 s( 99.95%)p 0.00( 0.05%)
9. (1.98271) BD ( 1) C 4 - C 5
( 50.77%) 0.7125* C 4 s( 37.60%)p 1.66( 62.37%)d 0.00( 0.03%)
( 49.23%) 0.7017* C 5 s( 32.51%)p 2.08( 67.45%)d 0.00( 0.05%)
10. (1.76874) BD ( 2) C 4 - C 5
( 48.12%) 0.6937* C 4 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
( 51.88%) 0.7202* C 5 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
11. (1.98568) BD ( 1) C 4 - H 9
( 59.32%) 0.7702* C 4 s( 26.85%)p 2.72( 73.10%)d 0.00( 0.05%)
( 40.68%) 0.6378* H 9 s( 99.95%)p 0.00( 0.05%)
12. (1.98422) BD ( 1) C 5 - H 10
( 59.41%) 0.7708* C 5 s( 25.40%)p 2.93( 74.55%)d 0.00( 0.05%)
( 40.59%) 0.6371* H 10 s( 99.95%)p 0.00( 0.05%)
13. (1.96998) BD ( 1) C 5 - B 12
( 66.70%) 0.8167* C 5 s( 42.00%)p 1.38( 57.99%)d 0.00( 0.01%)
( 33.30%) 0.5771* B 12 s( 33.40%)p 1.99( 66.53%)d 0.00( 0.08%)
14. (1.98608) BD ( 1) H 11 - B 12
( 55.09%) 0.7422* H 11 s( 99.97%)p 0.00( 0.03%)
( 44.91%) 0.6702* B 12 s( 33.17%)p 2.01( 66.78%)d 0.00( 0.06%)
15. (1.99902) CR ( 1) C 1 s(100.00%)p 0.00( 0.00%)
16. (1.99910) CR ( 1) C 2 s(100.00%)p 0.00( 0.00%)
17. (1.99907) CR ( 1) C 3 s(100.00%)p 0.00( 0.00%)
18. (1.99910) CR ( 1) C 4 s(100.00%)p 0.00( 0.00%)
19. (1.99902) CR ( 1) C 5 s(100.00%)p 0.00( 0.00%)
20. (1.99907) CR ( 1) B 12 s(100.00%)p 0.00( 0.00%)
It can be seen from above that C atoms are sp2 hybridised except for the C atoms bonded to B, which has 42% s character and 58% p character. the larger degree of s character may be due to the electron donating effect of boron which causes a greater electron density at the adjacent C atoms; s orbitals are less diffuse than p orbitals corresponding to a greater electron density nearer the nucleus at those C atoms. The boron atom has 33% s character and 67% p character indicating an sp2 hybridiastion.
Natural Bond Orbitals (Summary):
NBO Occupancy Energy
====================================================================================
Molecular unit 1 (C5H6B)
1. BD ( 1) C 1 - C 2 1.98271 -0.46501
2. BD ( 2) C 1 - C 2 1.76873 -0.02907
3. BD ( 1) C 1 - H 6 1.98422 -0.28859
4. BD ( 1) C 1 - B 12 1.96998 -0.31756
5. BD ( 1) C 2 - C 3 1.97969 -0.46986
6. BD ( 1) C 2 - H 7 1.98568 -0.31398
7. BD ( 1) C 3 - C 4 1.97969 -0.46986
8. BD ( 1) C 3 - H 8 1.98505 -0.31755
9. BD ( 1) C 4 - C 5 1.98271 -0.46502
10. BD ( 2) C 4 - C 5 1.76874 -0.02907
11. BD ( 1) C 4 - H 9 1.98568 -0.31398
12. BD ( 1) C 5 - H 10 1.98422 -0.28859
13. BD ( 1) C 5 - B 12 1.96998 -0.31756
14. BD ( 1) H 11 - B 12 1.98608 -0.17257
15. CR ( 1) C 1 1.99902 -9.79408
16. CR ( 1) C 2 1.99910 -9.83477)
17. CR ( 1) C 3 1.99907 -9.82821
18. CR ( 1) C 4 1.99910 -9.83477
19. CR ( 1) C 5 1.99902 -9.79408
20. CR ( 1) B 12 1.99907 -6.36952
21. LP ( 1) C 3 1.14703 0.09689
22. LP*( 1) B 12 0.57243 0.22263)
It can be seen that C-C bonds have an energy of -0.47, C-H bonds -0.31, C-B bonds -0.32 and B-H bonds -0.17. the C-B bond is much more stable than the B-H bond due to better orbital overlap between B and C, being in the same period, and C being better able to act as a Lewis base, being of lower electronegativity than H. Boron being electron deficient is a Lewis acid and is less well stabilised by H.
Range is from -1.000 - +1.000.
The boron atom is relatively positively charged and the carbon atoms adjacent to it display a higher electron density; this is due to the electropositive boron pushing electron density away from it towards the carbon atoms. The carbon in the para position is more negative than the 2 meta Cs indicating some degree of electron donating effect from boron across the ring. The H atom bonded to boron is also relatively positively charged compared to the other H atoms which are bonded to carbon in the molecule; boron is more electropositive than carbon and so this is to be expected.
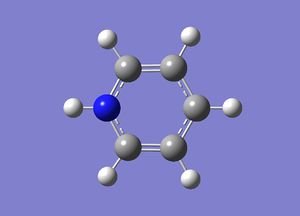
Pyridinium
The pyridinium ion is a benzene analogue with one C-H unit replaced by N-H. To be isoelectronic with benzene, a +1 charge is applied to the molecule.
Optimisation
Published in D-space: [11]
Item Value Threshold Converged?
Maximum Force 0.000064 0.000450 YES
RMS Force 0.000023 0.000300 YES
Maximum Displacement 0.000704 0.001800 YES
RMS Displacement 0.000174 0.001200 YES
Predicted change in Energy=-6.897392D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3839 -DE/DX = 0.0 !
! R2 R(1,6) 1.0832 -DE/DX = 0.0 !
! R3 R(1,12) 1.3523 -DE/DX = 0.0001 !
! R4 R(2,3) 1.3988 -DE/DX = 0.0 !
! R5 R(2,7) 1.0835 -DE/DX = 0.0 !
! R6 R(3,4) 1.3988 -DE/DX = 0.0 !
! R7 R(3,8) 1.0852 -DE/DX = 0.0 !
! R8 R(4,5) 1.3838 -DE/DX = 0.0 !
! R9 R(4,9) 1.0835 -DE/DX = 0.0 !
! R10 R(5,10) 1.0832 -DE/DX = 0.0 !
! R11 R(5,12) 1.3524 -DE/DX = 0.0 !
! R12 R(11,12) 1.0169 -DE/DX = 0.0 !
! A1 A(2,1,6) 123.9297 -DE/DX = 0.0 !
! A2 A(2,1,12) 119.2362 -DE/DX = 0.0 !
! A3 A(6,1,12) 116.8341 -DE/DX = 0.0 !
! A4 A(1,2,3) 119.082 -DE/DX = 0.0 !
! A5 A(1,2,7) 119.4193 -DE/DX = 0.0001 !
! A6 A(3,2,7) 121.4987 -DE/DX = -0.0001 !
! A7 A(2,3,4) 120.0549 -DE/DX = 0.0 !
! A8 A(2,3,8) 119.974 -DE/DX = 0.0 !
! A9 A(4,3,8) 119.9711 -DE/DX = 0.0 !
! A10 A(3,4,5) 119.0826 -DE/DX = 0.0 !
! A11 A(3,4,9) 121.4958 -DE/DX = -0.0001 !
! A12 A(5,4,9) 119.4215 -DE/DX = 0.0 !
! A13 A(4,5,10) 123.9324 -DE/DX = 0.0 !
! A14 A(4,5,12) 119.2354 -DE/DX = 0.0 !
! A15 A(10,5,12) 116.8322 -DE/DX = 0.0 !
! A16 A(1,12,5) 123.3087 -DE/DX = 0.0 !
! A17 A(1,12,11) 118.3463 -DE/DX = 0.0 !
! A18 A(5,12,11) 118.345 -DE/DX = 0.0 !
! D1 D(6,1,2,3) 180.0007 -DE/DX = 0.0 !
! D2 D(6,1,2,7) 0.0001 -DE/DX = 0.0 !
! D3 D(12,1,2,3) -0.0001 -DE/DX = 0.0 !
! D4 D(12,1,2,7) -180.0006 -DE/DX = 0.0 !
! D5 D(2,1,12,5) 0.0018 -DE/DX = 0.0 !
! D6 D(2,1,12,11) 180.0005 -DE/DX = 0.0 !
! D7 D(6,1,12,5) 180.0011 -DE/DX = 0.0 !
! D8 D(6,1,12,11) -0.0002 -DE/DX = 0.0 !
! D9 D(1,2,3,4) -0.0006 -DE/DX = 0.0 !
! D10 D(1,2,3,8) -179.9998 -DE/DX = 0.0 !
! D11 D(7,2,3,4) -180.0001 -DE/DX = 0.0 !
! D12 D(7,2,3,8) 0.0008 -DE/DX = 0.0 !
! D13 D(2,3,4,5) -0.0003 -DE/DX = 0.0 !
! D14 D(2,3,4,9) -179.9997 -DE/DX = 0.0 !
! D15 D(8,3,4,5) -180.0011 -DE/DX = 0.0 !
! D16 D(8,3,4,9) -0.0005 -DE/DX = 0.0 !
! D17 D(3,4,5,10) -180.0005 -DE/DX = 0.0 !
! D18 D(3,4,5,12) 0.002 -DE/DX = 0.0 !
! D19 D(9,4,5,10) -0.0011 -DE/DX = 0.0 !
! D20 D(9,4,5,12) 180.0013 -DE/DX = 0.0 !
! D21 D(4,5,12,1) -0.0028 -DE/DX = 0.0 !
! D22 D(4,5,12,11) -180.0014 -DE/DX = 0.0 !
! D23 D(10,5,12,1) -180.0005 -DE/DX = 0.0 !
! D24 D(10,5,12,11) 0.0009 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Frequency
Published in D-space: [12]
Item Value Threshold Converged?
Maximum Force 0.000004 0.000015 YES
RMS Force 0.000001 0.000010 YES
Maximum Displacement 0.000027 0.000060 YES
RMS Displacement 0.000007 0.000040 YES
Predicted change in Energy=-1.150410D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3838 -DE/DX = 0.0 !
! R2 R(1,6) 1.0832 -DE/DX = 0.0 !
! R3 R(1,12) 1.3524 -DE/DX = 0.0 !
! R4 R(2,3) 1.3987 -DE/DX = 0.0 !
! R5 R(2,7) 1.0835 -DE/DX = 0.0 !
! R6 R(3,4) 1.3987 -DE/DX = 0.0 !
! R7 R(3,8) 1.0852 -DE/DX = 0.0 !
! R8 R(4,5) 1.3838 -DE/DX = 0.0 !
! R9 R(4,9) 1.0835 -DE/DX = 0.0 !
! R10 R(5,10) 1.0832 -DE/DX = 0.0 !
! R11 R(5,12) 1.3524 -DE/DX = 0.0 !
! R12 R(11,12) 1.0169 -DE/DX = 0.0 !
! A1 A(2,1,6) 123.9398 -DE/DX = 0.0 !
! A2 A(2,1,12) 119.2367 -DE/DX = 0.0 !
! A3 A(6,1,12) 116.8235 -DE/DX = 0.0 !
! A4 A(1,2,3) 119.0803 -DE/DX = 0.0 !
! A5 A(1,2,7) 119.445 -DE/DX = 0.0 !
! A6 A(3,2,7) 121.4747 -DE/DX = 0.0 !
! A7 A(2,3,4) 120.0614 -DE/DX = 0.0 !
! A8 A(2,3,8) 119.9695 -DE/DX = 0.0 !
! A9 A(4,3,8) 119.9691 -DE/DX = 0.0 !
! A10 A(3,4,5) 119.0804 -DE/DX = 0.0 !
! A11 A(3,4,9) 121.4743 -DE/DX = 0.0 !
! A12 A(5,4,9) 119.4453 -DE/DX = 0.0 !
! A13 A(4,5,10) 123.9402 -DE/DX = 0.0 !
! A14 A(4,5,12) 119.2366 -DE/DX = 0.0 !
! A15 A(10,5,12) 116.8232 -DE/DX = 0.0 !
! A16 A(1,12,5) 123.3046 -DE/DX = 0.0 !
! A17 A(1,12,11) 118.3478 -DE/DX = 0.0 !
! A18 A(5,12,11) 118.3476 -DE/DX = 0.0 !
! D1 D(6,1,2,3) 180.0001 -DE/DX = 0.0 !
! D2 D(6,1,2,7) 0.0 -DE/DX = 0.0 !
! D3 D(12,1,2,3) 0.0 -DE/DX = 0.0 !
! D4 D(12,1,2,7) 180.0 -DE/DX = 0.0 !
! D5 D(2,1,12,5) 0.0001 -DE/DX = 0.0 !
! D6 D(2,1,12,11) -180.0 -DE/DX = 0.0 !
! D7 D(6,1,12,5) -180.0 -DE/DX = 0.0 !
! D8 D(6,1,12,11) 0.0 -DE/DX = 0.0 !
! D9 D(1,2,3,4) 0.0 -DE/DX = 0.0 !
! D10 D(1,2,3,8) -180.0 -DE/DX = 0.0 !
! D11 D(7,2,3,4) 180.0 -DE/DX = 0.0 !
! D12 D(7,2,3,8) 0.0 -DE/DX = 0.0 !
! D13 D(2,3,4,5) 0.0 -DE/DX = 0.0 !
! D14 D(2,3,4,9) -180.0001 -DE/DX = 0.0 !
! D15 D(8,3,4,5) -180.0 -DE/DX = 0.0 !
! D16 D(8,3,4,9) -0.0001 -DE/DX = 0.0 !
! D17 D(3,4,5,10) 179.9999 -DE/DX = 0.0 !
! D18 D(3,4,5,12) 0.0001 -DE/DX = 0.0 !
! D19 D(9,4,5,10) 0.0 -DE/DX = 0.0 !
! D20 D(9,4,5,12) 180.0001 -DE/DX = 0.0 !
! D21 D(4,5,12,1) -0.0001 -DE/DX = 0.0 !
! D22 D(4,5,12,11) -180.0001 -DE/DX = 0.0 !
! D23 D(10,5,12,1) 180.0 -DE/DX = 0.0 !
! D24 D(10,5,12,11) 0.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
All low frequencies were in a narrow range and there were no positive values.
Low frequencies --- -9.4128 -3.1616 -0.0009 -0.0009 -0.0008 1.0144 Low frequencies --- 391.8999 404.3405 620.2002
MO Analysis
Published in D-space: [13]
| MO | Image | MO | Image | MO | Image | MO | Image | MO | Image | MO | Image |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 7 | 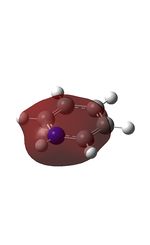 |
10 | 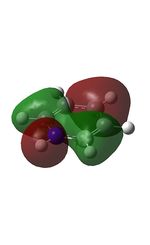 |
13 |  |
16 | 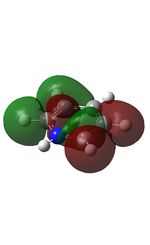 |
19 | 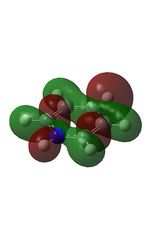 |
22 | 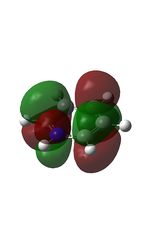
|
| 8 | 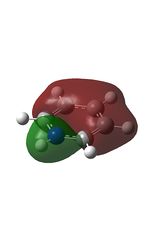 |
11 | 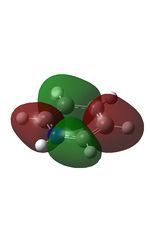 |
14 | 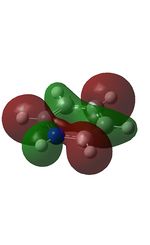 |
17 | 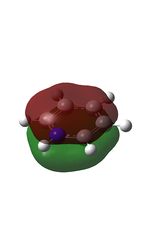 |
20 | 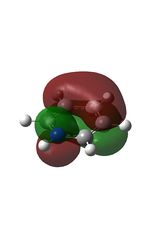 |
23 | 
|
| 9 |  |
12 |  |
15 |  |
18 |  |
21 | 
|
NBO Analysis
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
C 1 0.07102 1.99918 3.91065 0.01916 5.92898
C 2 -0.24106 1.99912 4.22862 0.01331 6.24106
C 3 -0.12240 1.99913 4.10941 0.01386 6.12240
C 4 -0.24106 1.99912 4.22862 0.01331 6.24106
C 5 0.07101 1.99918 3.91065 0.01916 5.92899
H 6 0.28493 0.00000 0.71397 0.00110 0.71507
H 7 0.29720 0.00000 0.70177 0.00103 0.70280
H 8 0.29169 0.00000 0.70718 0.00113 0.70831
H 9 0.29720 0.00000 0.70177 0.00103 0.70280
H 10 0.28493 0.00000 0.71397 0.00110 0.71507
H 11 0.48278 0.00000 0.51476 0.00246 0.51722
N 12 -0.47623 1.99937 5.46757 0.00929 7.47623
=======================================================================
* Total * 1.00000 11.99510 29.90895 0.09595 42.00000
Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98297) BD ( 1) C 1 - C 2
( 50.42%) 0.7100* C 1 s( 38.50%)p 1.60( 61.46%)d 0.00( 0.04%)
( 49.58%) 0.7042* C 2 s( 33.48%)p 1.99( 66.48%)d 0.00( 0.05%)
2. (1.98154) BD ( 1) C 1 - H 6
( 64.26%) 0.8016* C 1 s( 33.44%)p 1.99( 66.52%)d 0.00( 0.04%)
( 35.74%) 0.5978* H 6 s( 99.94%)p 0.00( 0.06%)
3. (1.98862) BD ( 1) C 1 - N 12
( 36.68%) 0.6057* C 1 s( 28.13%)p 2.55( 71.74%)d 0.00( 0.13%)
( 63.32%) 0.7957* N 12 s( 36.56%)p 1.73( 63.41%)d 0.00( 0.03%)
4. (1.82446) BD ( 2) C 1 - N 12
( 28.54%) 0.5343* C 1 s( 0.00%)p 1.00( 99.83%)d 0.00( 0.17%)
( 71.46%) 0.8453* N 12 s( 0.00%)p 1.00( 99.98%)d 0.00( 0.02%)
5. (1.98249) BD ( 1) C 2 - C 3
( 50.26%) 0.7089* C 2 s( 34.72%)p 1.88( 65.23%)d 0.00( 0.04%)
( 49.74%) 0.7053* C 3 s( 34.45%)p 1.90( 65.51%)d 0.00( 0.04%)
6. (1.54876) BD ( 2) C 2 - C 3
( 54.27%) 0.7367* C 2 s( 0.00%)p 1.00( 99.94%)d 0.00( 0.06%)
( 45.73%) 0.6762* C 3 s( 0.00%)p 1.00( 99.93%)d 0.00( 0.07%)
7. (1.97822) BD ( 1) C 2 - H 7
( 64.83%) 0.8052* C 2 s( 31.78%)p 2.15( 68.19%)d 0.00( 0.03%)
( 35.17%) 0.5930* H 7 s( 99.94%)p 0.00( 0.06%)
8. (1.98249) BD ( 1) C 3 - C 4
( 49.74%) 0.7053* C 3 s( 34.45%)p 1.90( 65.51%)d 0.00( 0.04%)
( 50.26%) 0.7089* C 4 s( 34.72%)p 1.88( 65.23%)d 0.00( 0.04%)
9. (1.98140) BD ( 1) C 3 - H 8
( 64.64%) 0.8040* C 3 s( 31.07%)p 2.22( 68.90%)d 0.00( 0.03%)
( 35.36%) 0.5947* H 8 s( 99.94%)p 0.00( 0.06%)
10. (1.98297) BD ( 1) C 4 - C 5
( 49.58%) 0.7042* C 4 s( 33.48%)p 1.99( 66.48%)d 0.00( 0.05%)
( 50.42%) 0.7100* C 5 s( 38.50%)p 1.60( 61.46%)d 0.00( 0.04%)
11. (1.61443) BD ( 2) C 4 - C 5
( 52.23%) 0.7227* C 4 s( 0.00%)p 1.00( 99.94%)d 0.00( 0.06%)
( 47.77%) 0.6912* C 5 s( 0.00%)p 1.00( 99.94%)d 0.00( 0.06%)
12. (1.97822) BD ( 1) C 4 - H 9
( 64.83%) 0.8052* C 4 s( 31.78%)p 2.15( 68.19%)d 0.00( 0.03%)
( 35.17%) 0.5930* H 9 s( 99.94%)p 0.00( 0.06%)
13. (1.98154) BD ( 1) C 5 - H 10
( 64.26%) 0.8016* C 5 s( 33.44%)p 1.99( 66.52%)d 0.00( 0.04%)
( 35.74%) 0.5978* H 10 s( 99.94%)p 0.00( 0.06%)
14. (1.98862) BD ( 1) C 5 - N 12
( 36.68%) 0.6057* C 5 s( 28.13%)p 2.55( 71.74%)d 0.00( 0.13%)
( 63.32%) 0.7957* N 12 s( 36.56%)p 1.73( 63.41%)d 0.00( 0.03%)
15. (1.98630) BD ( 1) H 11 - N 12
( 25.41%) 0.5041* H 11 s( 99.88%)p 0.00( 0.12%)
( 74.59%) 0.8637* N 12 s( 26.82%)p 2.73( 73.15%)d 0.00( 0.02%)
16. (1.99918) CR ( 1) C 1 s(100.00%)p 0.00( 0.00%)
17. (1.99913) CR ( 1) C 2 s(100.00%)p 0.00( 0.00%)
18. (1.99914) CR ( 1) C 3 s(100.00%)p 0.00( 0.00%)
19. (1.99913) CR ( 1) C 4 s(100.00%)p 0.00( 0.00%)
20. (1.99918) CR ( 1) C 5 s(100.00%)p 0.00( 0.00%)
21. (1.99937) CR ( 1) N 12 s(100.00%)p 0.00( 0.00%)
N and Care sp2 hybridised.
Range is from -1.000 - +1.000.
The electron density is greatest at the N atom; this is to be expected given the electronegative nature of nitrogen. The carbon atoms adjacent to N are relatively positively charged compared to the other C atoms (+0.071 vs. -0.241); this is due to the electron withdrawing effect of N.
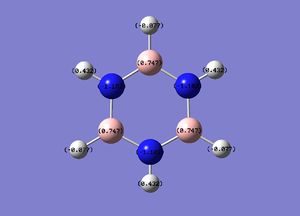
Borazine
Optimisation
Published in D-space: [14]
The N-B bond length is intermediate between a single B-N bond (0.151Å) and a double B=N bond (0.131Å), indicating delocalisation of the nitrogen lone pair electrons.
Item Value Threshold Converged?
Maximum Force 0.000086 0.000450 YES
RMS Force 0.000033 0.000300 YES
Maximum Displacement 0.000252 0.001800 YES
RMS Displacement 0.000077 0.001200 YES
Predicted change in Energy=-9.332344D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,8) 1.1949 -DE/DX = 0.0001 !
! R2 R(2,12) 1.0097 -DE/DX = 0.0 !
! R3 R(3,9) 1.1949 -DE/DX = 0.0001 !
! R4 R(4,11) 1.0097 -DE/DX = 0.0 !
! R5 R(5,7) 1.1949 -DE/DX = 0.0001 !
! R6 R(6,10) 1.0097 -DE/DX = 0.0 !
! R7 R(7,10) 1.4307 -DE/DX = -0.0001 !
! R8 R(7,11) 1.4307 -DE/DX = -0.0001 !
! R9 R(8,10) 1.4306 -DE/DX = -0.0001 !
! R10 R(8,12) 1.4307 -DE/DX = -0.0001 !
! R11 R(9,11) 1.4307 -DE/DX = -0.0001 !
! R12 R(9,12) 1.4307 -DE/DX = -0.0001 !
! A1 A(5,7,10) 121.437 -DE/DX = 0.0 !
! A2 A(5,7,11) 121.4399 -DE/DX = 0.0 !
! A3 A(10,7,11) 117.123 -DE/DX = 0.0 !
! A4 A(1,8,10) 121.4407 -DE/DX = 0.0 !
! A5 A(1,8,12) 121.4364 -DE/DX = 0.0 !
! A6 A(10,8,12) 117.1229 -DE/DX = 0.0 !
! A7 A(3,9,11) 121.4374 -DE/DX = 0.0 !
! A8 A(3,9,12) 121.4415 -DE/DX = 0.0 !
! A9 A(11,9,12) 117.1211 -DE/DX = 0.0 !
! A10 A(6,10,7) 118.5602 -DE/DX = 0.0 !
! A11 A(6,10,8) 118.5628 -DE/DX = 0.0 !
! A12 A(7,10,8) 122.877 -DE/DX = 0.0 !
! A13 A(4,11,7) 118.5641 -DE/DX = 0.0 !
! A14 A(4,11,9) 118.5579 -DE/DX = 0.0 !
! A15 A(7,11,9) 122.878 -DE/DX = 0.0 !
! A16 A(2,12,8) 118.5609 -DE/DX = 0.0 !
! A17 A(2,12,9) 118.561 -DE/DX = 0.0 !
! A18 A(8,12,9) 122.8781 -DE/DX = 0.0 !
! D1 D(5,7,10,6) 0.0003 -DE/DX = 0.0 !
! D2 D(5,7,10,8) 179.997 -DE/DX = 0.0 !
! D3 D(11,7,10,6) -179.9999 -DE/DX = 0.0 !
! D4 D(11,7,10,8) -0.0032 -DE/DX = 0.0 !
! D5 D(5,7,11,4) 0.0036 -DE/DX = 0.0 !
! D6 D(5,7,11,9) -179.9997 -DE/DX = 0.0 !
! D7 D(10,7,11,4) -179.9962 -DE/DX = 0.0 !
! D8 D(10,7,11,9) 0.0005 -DE/DX = 0.0 !
! D9 D(1,8,10,6) -0.0025 -DE/DX = 0.0 !
! D10 D(1,8,10,7) -179.9992 -DE/DX = 0.0 !
! D11 D(12,8,10,6) 179.9975 -DE/DX = 0.0 !
! D12 D(12,8,10,7) 0.0008 -DE/DX = 0.0 !
! D13 D(1,8,12,2) -0.0057 -DE/DX = 0.0 !
! D14 D(1,8,12,9) -179.9955 -DE/DX = 0.0 !
! D15 D(10,8,12,2) 179.9944 -DE/DX = 0.0 !
! D16 D(10,8,12,9) 0.0045 -DE/DX = 0.0 !
! D17 D(3,9,11,4) 0.0007 -DE/DX = 0.0 !
! D18 D(3,9,11,7) -179.996 -DE/DX = 0.0 !
! D19 D(12,9,11,4) -179.9989 -DE/DX = 0.0 !
! D20 D(12,9,11,7) 0.0044 -DE/DX = 0.0 !
! D21 D(3,9,12,2) 0.0035 -DE/DX = 0.0 !
! D22 D(3,9,12,8) 179.9933 -DE/DX = 0.0 !
! D23 D(11,9,12,2) -179.9968 -DE/DX = 0.0 !
! D24 D(11,9,12,8) -0.007 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Frequency
Published in D-space [15]
Item Value Threshold Converged?
Maximum Force 0.000010 0.000015 YES
RMS Force 0.000005 0.000010 YES
Maximum Displacement 0.000055 0.000060 YES
RMS Displacement 0.000024 0.000040 YES
Predicted change in Energy=-3.593381D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,8) 1.1951 -DE/DX = 0.0 !
! R2 R(2,12) 1.0097 -DE/DX = 0.0 !
! R3 R(3,9) 1.1951 -DE/DX = 0.0 !
! R4 R(4,11) 1.0097 -DE/DX = 0.0 !
! R5 R(5,7) 1.1951 -DE/DX = 0.0 !
! R6 R(6,10) 1.0097 -DE/DX = 0.0 !
! R7 R(7,10) 1.4306 -DE/DX = 0.0 !
! R8 R(7,11) 1.4306 -DE/DX = 0.0 !
! R9 R(8,10) 1.4306 -DE/DX = 0.0 !
! R10 R(8,12) 1.4306 -DE/DX = 0.0 !
! R11 R(9,11) 1.4306 -DE/DX = 0.0 !
! R12 R(9,12) 1.4306 -DE/DX = 0.0 !
! A1 A(5,7,10) 121.4352 -DE/DX = 0.0 !
! A2 A(5,7,11) 121.4365 -DE/DX = 0.0 !
! A3 A(10,7,11) 117.1283 -DE/DX = 0.0 !
! A4 A(1,8,10) 121.4373 -DE/DX = 0.0 !
! A5 A(1,8,12) 121.4344 -DE/DX = 0.0 !
! A6 A(10,8,12) 117.1283 -DE/DX = 0.0 !
! A7 A(3,9,11) 121.4348 -DE/DX = 0.0 !
! A8 A(3,9,12) 121.4369 -DE/DX = 0.0 !
! A9 A(11,9,12) 117.1283 -DE/DX = 0.0 !
! A10 A(6,10,7) 118.563 -DE/DX = 0.0 !
! A11 A(6,10,8) 118.5646 -DE/DX = 0.0 !
! A12 A(7,10,8) 122.8724 -DE/DX = 0.0 !
! A13 A(4,11,7) 118.5653 -DE/DX = 0.0 !
! A14 A(4,11,9) 118.5634 -DE/DX = 0.0 !
! A15 A(7,11,9) 122.8713 -DE/DX = 0.0 !
! A16 A(2,12,8) 118.5636 -DE/DX = 0.0 !
! A17 A(2,12,9) 118.565 -DE/DX = 0.0 !
! A18 A(8,12,9) 122.8714 -DE/DX = 0.0 !
! D1 D(5,7,10,6) -0.0001 -DE/DX = 0.0 !
! D2 D(5,7,10,8) 179.9987 -DE/DX = 0.0 !
! D3 D(11,7,10,6) 180.0004 -DE/DX = 0.0 !
! D4 D(11,7,10,8) -0.0008 -DE/DX = 0.0 !
! D5 D(5,7,11,4) 0.0018 -DE/DX = 0.0 !
! D6 D(5,7,11,9) -179.9996 -DE/DX = 0.0 !
! D7 D(10,7,11,4) 180.0013 -DE/DX = 0.0 !
! D8 D(10,7,11,9) -0.0001 -DE/DX = 0.0 !
! D9 D(1,8,10,6) -0.0012 -DE/DX = 0.0 !
! D10 D(1,8,10,7) 180.0001 -DE/DX = 0.0 !
! D11 D(12,8,10,6) 179.9987 -DE/DX = 0.0 !
! D12 D(12,8,10,7) -0.0001 -DE/DX = 0.0 !
! D13 D(1,8,12,2) -0.0024 -DE/DX = 0.0 !
! D14 D(1,8,12,9) -179.9982 -DE/DX = 0.0 !
! D15 D(10,8,12,2) 179.9978 -DE/DX = 0.0 !
! D16 D(10,8,12,9) 0.0019 -DE/DX = 0.0 !
! D17 D(3,9,11,4) 0.0008 -DE/DX = 0.0 !
! D18 D(3,9,11,7) 180.0021 -DE/DX = 0.0 !
! D19 D(12,9,11,4) -179.9996 -DE/DX = 0.0 !
! D20 D(12,9,11,7) 0.0018 -DE/DX = 0.0 !
! D21 D(3,9,12,2) 0.0011 -DE/DX = 0.0 !
! D22 D(3,9,12,8) 179.9969 -DE/DX = 0.0 !
! D23 D(11,9,12,2) -179.9986 -DE/DX = 0.0 !
! D24 D(11,9,12,8) -0.0027 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Low frequencies --- -3.9251 -3.1910 -0.0010 -0.0009 0.0009 4.6183 Low frequencies --- 289.7172 289.8015 404.5572
MO Analysis
Published in D-space: [16]
| MO | Image | MO | Image | MO | Image | MO | Image | MO | Image | MO | Image |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 7 |  |
10 |  |
13 |  |
16 |  |
19 |  |
22 | 
|
| 8 |  |
11 |  |
14 | 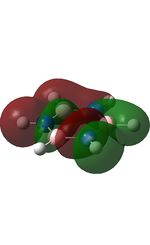 |
17 | 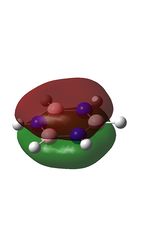 |
20 | 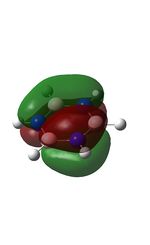 |
23 | 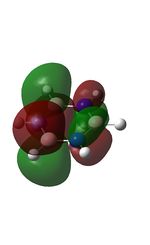
|
| 9 | 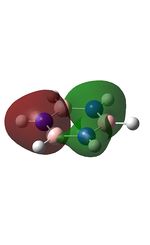 |
12 | 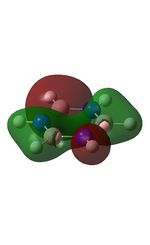 |
15 |  |
18 |  |
21 | 
|
NBO Analysis
From NBO analysis it can be seen that all B atoms are identical and all N atoms are identical in their electron density. Hydrogens bonded to nitrogen are relatively more positive than H atoms bonded to boron; this is to be expected due to the electronegative nature of nitrogen withdrawing local electron density and the electropositive nature of boron increasing electron density.
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
H 1 -0.07654 0.00000 1.07585 0.00069 1.07654
H 2 0.43200 0.00000 0.56572 0.00228 0.56800
H 3 -0.07653 0.00000 1.07584 0.00069 1.07653
H 4 0.43200 0.00000 0.56572 0.00228 0.56800
H 5 -0.07654 0.00000 1.07585 0.00069 1.07654
H 6 0.43200 0.00000 0.56572 0.00228 0.56800
B 7 0.74695 1.99917 2.23867 0.01521 4.25305
B 8 0.74695 1.99917 2.23867 0.01521 4.25305
B 9 0.74692 1.99917 2.23870 0.01521 4.25308
N 10 -1.10240 1.99943 6.09819 0.00478 8.10240
N 11 -1.10240 1.99943 6.09819 0.00478 8.10240
N 12 -1.10240 1.99943 6.09820 0.00478 8.10240
=======================================================================
* Total * 0.00000 11.99579 29.93531 0.06890 42.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98670) BD ( 1) H 1 - B 8
( 54.03%) 0.7351* H 1 s( 99.96%)p 0.00( 0.04%)
( 45.97%) 0.6780* B 8 s( 37.47%)p 1.67( 62.47%)d 0.00( 0.07%)
2. (1.98494) BD ( 1) H 2 - N 12
( 28.08%) 0.5299* H 2 s( 99.91%)p 0.00( 0.09%)
( 71.92%) 0.8481* N 12 s( 22.82%)p 3.38( 77.15%)d 0.00( 0.03%)
3. (1.98670) BD ( 1) H 3 - B 9
( 54.03%) 0.7351* H 3 s( 99.96%)p 0.00( 0.04%)
( 45.97%) 0.6780* B 9 s( 37.47%)p 1.67( 62.47%)d 0.00( 0.07%)
4. (1.98494) BD ( 1) H 4 - N 11
( 28.08%) 0.5299* H 4 s( 99.91%)p 0.00( 0.09%)
( 71.92%) 0.8481* N 11 s( 22.82%)p 3.38( 77.15%)d 0.00( 0.03%)
5. (1.98670) BD ( 1) H 5 - B 7
( 54.03%) 0.7351* H 5 s( 99.96%)p 0.00( 0.04%)
( 45.97%) 0.6780* B 7 s( 37.47%)p 1.67( 62.47%)d 0.00( 0.07%)
6. (1.98494) BD ( 1) H 6 - N 10
( 28.08%) 0.5299* H 6 s( 99.91%)p 0.00( 0.09%)
( 71.92%) 0.8481* N 10 s( 22.82%)p 3.38( 77.15%)d 0.00( 0.03%)
7. (1.98437) BD ( 1) B 7 - N 10
( 23.53%) 0.4851* B 7 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
( 76.47%) 0.8745* N 10 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
8. (1.98437) BD ( 1) B 7 - N 11
( 23.53%) 0.4851* B 7 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
( 76.47%) 0.8745* N 11 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
9. (1.82089) BD ( 2) B 7 - N 11
( 11.79%) 0.3433* B 7 s( 0.00%)p 1.00( 99.62%)d 0.00( 0.38%)
( 88.21%) 0.9392* N 11 s( 0.00%)p 1.00(100.00%)d 0.00( 0.00%)
10. (1.98437) BD ( 1) B 8 - N 10
( 23.53%) 0.4851* B 8 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
( 76.47%) 0.8745* N 10 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
11. (1.82089) BD ( 2) B 8 - N 10
( 11.79%) 0.3433* B 8 s( 0.00%)p 1.00( 99.62%)d 0.00( 0.38%)
( 88.21%) 0.9392* N 10 s( 0.00%)p 1.00(100.00%)d 0.00( 0.00%)
12. (1.98437) BD ( 1) B 8 - N 12
( 23.53%) 0.4851* B 8 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
( 76.47%) 0.8745* N 12 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
13. (1.98437) BD ( 1) B 9 - N 11
( 23.53%) 0.4851* B 9 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
( 76.47%) 0.8745* N 11 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
14. (1.98437) BD ( 1) B 9 - N 12
( 23.53%) 0.4851* B 9 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
( 76.47%) 0.8745* N 12 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
15. (1.82090) BD ( 2) B 9 - N 12
( 11.79%) 0.3433* B 9 s( 0.00%)p 1.00( 99.62%)d 0.00( 0.38%)
( 88.21%) 0.9392* N 12 s( 0.00%)p 1.00(100.00%)d 0.00( 0.00%)
16. (1.99917) CR ( 1) B 7 s(100.00%)p 0.00( 0.00%)
17. (1.99917) CR ( 1) B 8 s(100.00%)p 0.00( 0.00%)
18. (1.99917) CR ( 1) B 9 s(100.00%)p 0.00( 0.00%)
19. (1.99943) CR ( 1) N 10 s(100.00%)p 0.00( 0.00%)
20. (1.99943) CR ( 1) N 11 s(100.00%)p 0.00( 0.00%)
21. (1.99943) CR ( 1) N 12 s(100.00%)p 0.00( 0.00%)
B is sp2 hybridised; N has greater s character than B (39% for N vs. 31% for B). N is more electronegative and the s orbital is less diffuse; N has a greater electron density closer to its nucleus.
Range is from -1.000 - +1.000.
Comparison of Charge Distribution
Below shows the series' charge distributions and totally bonding pi orbital.
| Benzene | Boratabenzene | Pyridinium | Borazine |
|---|---|---|---|
 |
 |
 |
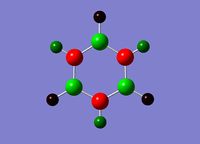
|
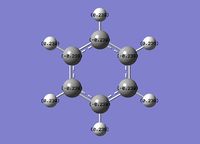 |
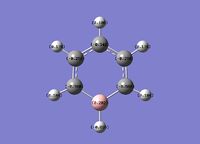 |
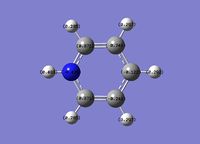 |
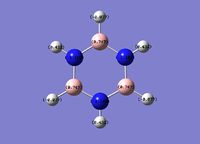
|
Nitrogen in borazine, bonded to 2 boron atoms, is more negatively charged (-1.107) than N in pyridinium bonded to two C atoms (-0.476). This is due to boron being more electropositive than carbon, thus pushing the electron density towards N.
The C atoms directly bonded to N in pyridinium are relatively more positively charged (+0.071) than the C atoms 2 bonds away from N (-0.241). The C atom in the para position is more positively charged than the meta Cs, indicating some degree of electron withdrawing effect of N across the diameter of the ring. The converse is true in boratabenzene, where the ortho Cs are the most negatively charged (-0.588) and the meta are the least negatively charged (-0.250); the para C has an intermediate charge of -0.340 indicating some degree of electron-donating behaviour from boron across the ring.
The H atoms then bonded to the Cs above display the converse effect - if C is relatively positive due to an electronegative substituent, the H bonded to it is correspondingly more negative and vice versa.
The H bonded directly to N in pyridinium is by far the most positive H atom in the molecule (+0.483), again due to the electron withdrawing effect of N. Conversely, the H bonded directly to B in boratabenzene is the most negative H atom (-0.096) due to the electron donating effect of B. Both of these effects are observed in borazine, where every H atom bonded to N is positive (+0.432) and every H bonded to B is negative (-0.077). All Hs have identical relative charges in benzene, as do all C atoms.
Comparison of MOs
| MO | Benzene | Boratabenzene | Pyridinium | Borazine |
|---|---|---|---|---|
| Pi totally bonding | 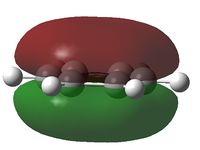 |
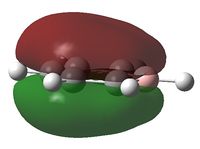 |
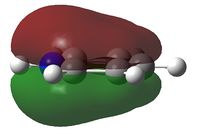 |
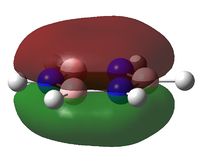
|
| LCAO | 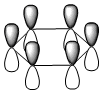 |
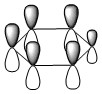 |
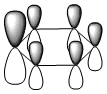 |
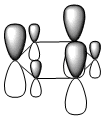 |
| Energy | -0.36 | -0.31 | -0.64 | -0.36 |
All four molecules have an delocalised electron ring above and below the plane of the molecule; this aromatic sextet is iconic of benzene. The lone pair of N in pyridine is not involved in the aromatic sextet and the pyridinium ion is aromatic; boratabenzene's charge of -1 means the pz orbital of B is filled and participates in the delocalised electron ring. All the totally pi bonding orbitals, as expected, have one nodal plane in the plane of the ring.
The lowest energy orbital is that of pyridinium. This is to be expected as the energy of the molecular orbital is largely determined by the energy of its atomic orbitals; pyridinium's N atom is more electronegative than carbon and lowers the energy of most of pyridine's MOs, whereas benzene contains only C (and H) and therefore has orbitals of higher energy. Boratabenzene, which has electropositive boron in place of nitrogen, is the highest energy. Borazine contains 3 electronegative N atoms but also 3 electropositive B atoms; overall the electronegativity is greatest in pyridinium. For all molecular orbitals, pyridine has the lowest energy and boratabenzene the highest, with benzene and borazine at an intermediate level.
The electronegativity of N can be seen in the MO with a larger electron density close to the N atom than the rest of the ring. Similarly, there is a smaller electron density near the B atom in boratabenzene relative to the rest of its ring. Benzene has a symmetrical electron distribution over the whole molecule showing that all C-H bonds in benzene are identical. Borazine too has a fairly evenly distributed electron distribution as the difference in electronegativity of the alternating N and B atoms averages out over the molecule. All these effects are seen in the LCAOs; the p orbital of N is much larger than those of C and the p orbital of B is smaller than those of C.
| MO | Benzene | Boratabenzene | Pyridinium | Borazine |
|---|---|---|---|---|
| Pi bonding HOMO-1 | 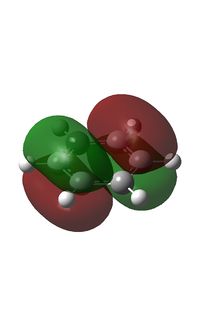 |
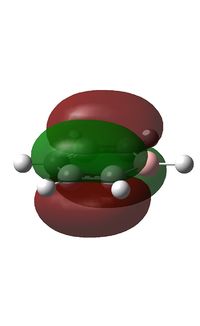 |
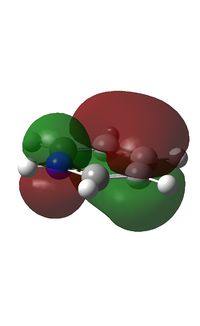 |
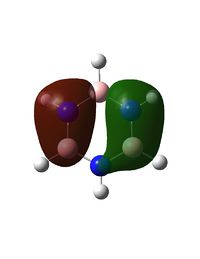
|
| LCAO | 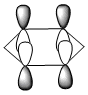 |
 |
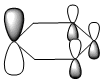 |
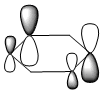 |
| Energy | -0.25 | -0.03 | -0.52 | -0.28 |
| MO | Benzene | Boratabenzene | Pyridinium | Borazine |
|---|---|---|---|---|
| Pi bonding HOMO | 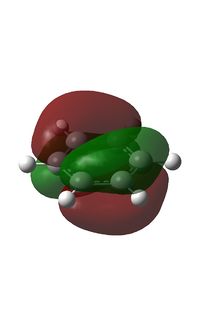 |
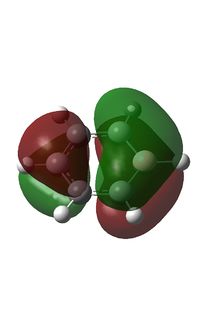 |
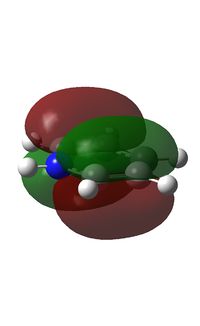 |
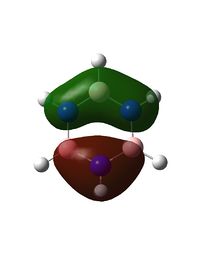
|
| LCAO | 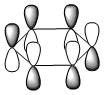 |
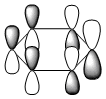 |
 |
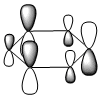 |
| Energy | -0.25 | +0.01 | -0.48 | -0.28 |
All molecules' HOMOs and HOMO-1s have a nodal plane in the plane of the ring and one nodal plane perpendicular to the plane of the ring. For benzene and borazine, the HOMO is doubly degenerate due to the symmetry of the molecule. This degeneracy is lost with pyridinium and boratabenzene, as the substitution of a carbon for an electronegative (electropositive) atom lowers (raises) the energy of the orbitals. Pyridinium's HOMO-1 orbital is slightly lower in energy than the HOMO as there is electron density present at the electronegative N atom, compared to its HOMO where there is a node at the N atom. From the LCAO of the HOMO-1, one can see the electron-withdrawing effect of N from the greater size of N's p orbital and the lack of electron density immediately next to the N atom.
The converse is true for boratabenzene's HOMO-1 - there is a node at the B atom on the HOMO-1 and electron density is present at the B atom on the HOMO. Boron being electropositive is destabilised by having a greater electron density. It can be seen that there is a greater electron density near the boron atom in the HOMO - this is unsurprising given its -1 charge. Due to boron's electron donating effects, the adjacent C atoms are more negatively charged and have a greater electron density than the rest of the Cs in the molecule, as evidenced previously in analysis of the charge distribution. This results in an asymmetrical electron distribution across the whole molecule; the orbitals on the B atom side are larger than the orbitals of the 3 C atoms.
This effect is also visible in the LCAOs of borazine. For the top HOMO, the orbital covering 2 N and 1 B atoms (green) is larger in size than the one covering 1 N and 2 B atoms (red), due to the electronegativity of N. In the HOMO, it can be seen that some of the electron density is drawn towards a N atom despite there being a nodal plane at the atom; this is not observed at the B atom of the nodal plane.
The effect of substitution on the overall MO diagram for these analogues can be rationalised by the relative energies of the AOs. Nitrogen being more electronegative than carbon is lower in energy and results in a lowering in energy of the MOs and a stabilisation of the pyridinium ion. Boron being less electronegative than carbon is higher in energy, raising the energy and destabilising the boratabenzene anion. The HOMO's energy is positive indicating the molecule is somewhat unstable though borabenzene in the free form with no Lewis base adduct has been isolated [4].
In borazine, N and B have very large electronegativity differences and therefore different AO energies. Boron has far larger orbital coefficients in the antibonding MOs and nitrogen larger in the bonding MOs. This results in an unequal pi electron distribution across the molecule, making borazine less aromatic than benzene.
References
- ↑ 1.0 1.1 J. Glaser and G. Johansson, Acta Chemica Scandinavica 1982, 36a, 125-135
- ↑ 2.0 2.1 M.R. Hartman, J.J. Rush, T.J. Udovic, R.C. Bowman and S.J. Hwang, Journal of Solid State Chemistry 2007, 180, 1298-1305
- ↑ 3.0 3.1 Yu. Kh. Shaulov; G. O. Shmyreva; V. S. Tubyanskaya, Zhurnal Fizicheskoi Khimii, 1966, 40,122-124
- ↑ 4.0 4.1 G. Maier, H.P. Reisenauer, J. Henkelmann, C. Kliche, Angew. Chem. Int. Ed. Engl., 27: 295–296.