Rep:Mod:blackmesa
Molecular Mechanics Techniques and Semi-Empirical Molecular Orbital Methods
Hydrogenation of Cyclopentadiene dimers
Cyclopentadiene readily undergoes a self Diels-Alder reaction, to give a dimer, at room temperature. As is the case with Diels-Alder reactions, an endo or exo product is possible from the reaction of cyclopentadiene. Modelling these two molecules in a molecular modelling program allows for a variety of information regarding the total energy of the molecules and how that can be attributed to different potential energy factors such as bond distances, angles, torsion angles and intramolecular interactions.
Cyclopentadiene Diels-Alder adducts
The endo adduct is the kinetic product of the Diels-Alder dimerisation of cyclopentadiene because it is higher in energy than the exo product but when the reacting cyclopentadiene molecules approach each other in the orientation necessary to give the endo product a secondary orbital interaction
| Molecular Property | Energy Contribution to Total Energy/kcal mol-1 |
|---|---|
| Bond Stretching | 1.3 |
| Bond Bending | 20.8 |
| Stretch-Bend | -0.8 |
| Torsions | 9.5 |
| Non 1,4 Van der Waals interactions | -1.5 |
| 1,4 Van der Waals interactions | 4.3 |
| Dipole-Dipole interactions | 0.4 |
| Total Energy | 34.0 |
 |

|
| Image of endo adduct of cyclopentadiene dimer | |
The Exo Adduct
| Molecular Property | Energy Contribution to Total Energy/kcal mol-1 |
|---|---|
| Bond Stretching | 1.3 |
| Bond Bending | 20.6 |
| Stretch-Bend | -0.8 |
| Torsions | 7.7 |
| Non 1,4 Van der Waals interactions | -1.4 |
| 1,4 Van der Waals interactions | 4.2 |
| Dipole-Dipole interactions | 0.4 |
| Total Energy | 31.9 |
 |

|
| Image of endo adduct of cyclopentadiene dimer | |
With this information the kinetic and thermodynamic products can be identified. The exo product is clearly lower in energy than the endo product and this is largely due to the difference in molecular strain. This is immediately obvious as the endo product is bent back on itself with the bond angle between carbon atoms C6, C5 and C8 being 117.9° in the endo structure and the equivalent angle in the exo structure being 114.3°, and therefore closer to the ideal tetrahedral geometry of 109.5 making the exo structure less strained. The endo adduct is the labelled the kinetic product as a secondary orbital interaction lowers the activation energy barrier. This occurs only for the endo adduct as the approaching molecules have to be better alligned on top of one another for the interaction to occur.
Hydrogenated Cyclopentadiene dimers
Subsequent reactions involving the two adducts can generate products that differ in energy, below is some data for two molecules that are formed from hydrogenating one of the two bonds in the cyclopentadiene endo adduct.
Hydrogenated Cyclopentadiene endo adduct (9,10 hydrogenation) no.3
| Molecular Property | Energy Contribution to Total Energy/kcal mol-1 |
|---|---|
| Bond Stretching | 1.3 |
| Bond Bending | 19.9 |
| Stretch-Bend | -0.8 |
| Torsions | 10.8 |
| Non 1,4 Van der Waals interactions | -1.2 |
| 1,4 Van der Waals interactions | 5.6 |
| Dipole-Dipole interactions | 0.2 |
| Total Energy | 35.7 |
Hydrogenated Cyclopentadiene endo adduct (1,2 hydrogenation) no.4
| Molecular Property | Energy Contribution to Total Energy/kcal mol-1 |
|---|---|
| Bond Stretching | 1.1 |
| Bond Bending | 14.5 |
| Stretch-Bend | -0.5 |
| Torsions | 12.5 |
| Non 1,4 Van der Waals interactions | -1.1 |
| 1,4 Van der Waals interactions | 4.5 |
| Dipole-Dipole interactions | 0.1 |
| Total Energy | 31.2 |
There isn't a huge disparity between the majority of the molecular properties that influence the total energy of these two molecules. The bond stretching, stretch-bending, non-1,4 Van der Waals interactions and dipole-dipole interactions are almost the same for both molecules. The largest differences are in the bond bending and bond torsions.
Taxol
Taxol: Molecule 9 (Upward pointing carbonyl)
| Molecular Property | Energy value /kcal/mol-1 |
|---|---|
| Bond Stretching | 6.4 |
| Bond Bending | 90.0 |
| Stretch-Bend | -0.4 |
| Torsion | 19.2 |
| Non-1,4 Van der Waals interaction | 2.7 |
| 1,4 Van der Waals interaction | 29.8 |
| Dipole-Dipole interaction | -2.8 |
| Total energy | 145.0 |
Taxol |
MMFF94 energy minimisation gave a final energy of 82.4 kcal/mol-1
Taxol: Moelcule 10 (downward pointing carbonyl group)
| Molecular Property | Energy value /kcal/mol-1 |
|---|---|
| Bond Stretching | 2.9 |
| Bond Bending | 16.8 |
| Stretch-Bend | -0.4 |
| Torsion | 20.2 |
| Non-1,4 Van der Waals interaction | 0.14 |
| 1,4 Van der Waals interaction | 14.3 |
| Dipole-Dipole interaction | -1.7 |
| Total energy | 53.2 |
An MMFF94 optimisation increased the total energy of the molecule to 82.3 kcal/mol-1
Investigations in to alkene strain energies show that typically a strained alkene will have more strain energy than its equivalent strained alkane. Alkene strain energies are calculated by subtracting the strain energy of the alkane from the strain energy of the alkene. This will usually give a positive energy, but there is a class of strained alkenes that give negative alkene strain energies labelled "hyperstable" alkenes. These types of compounds are often characterised by having a lower than predicted enthalpy of hydrogenation (ΔHhyd in comparison with similar acyclic alkenes with the same level of substitution[1] [2]. A common example of such alkenes are twisted, bridgehead junction, tri-substituted alkenes. Because there is less strain present in the alkene than in the equivalent alkene, there is therefore a preference for the double bond not react for the molecule to become saturated and so there must be some inherent thermodynamic stability to the bridgehead alkene.
Running an MM2 forcefield energy minimisation for the hydrogen equivalent of molecule 9 gives a total energy of 71.5 kcal/mol-1, with the increase in energy corresponding to increases in bond torsions and bond bending which would support the theory that bridgehead junction alkenes are less strained than their equivalent alkanes. Full energy contribution breakdown below.
Reaction of dichlorocarbene with bridged-bicyclic compound
When overlaying the two structures, whose geometry was optimised by different methods, it was noticed that poorest overlap occurred furthest away from the three atoms chosen to be superimposed. In the first example, the three atoms chosen were all part of the same ring in this fused, bicyclic structure. The atoms chosen were C1, C5 and C6, an sp2 hybridised carbon, the carbon atom at the closest bridgehead junction and the intervening carbon atom between the first two, respectively.
LUMO+2 energy = +5.77 eV LUMO+1 energy = +5.31 eV LUMO energy = +4.51 eV HOMO energy = -8.68 eV HOMO-1 energy = -9.44 eV HOMO-2 energy = -10.53 eV
Symmetry of molecular orbitals account for the symmetry plane that is perpendicular to the ring bridge.
The molecular vibrations for the compound were determined through a frequency analysis in gaussian. This was performed on an optimised molecule, with enforced Cs symmetry, that was imported in to gaussian from ChemBio3D as a gaussian input file. The input was based on the MOPAC optimised geometry. All calculations were performed on the HPC.
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G d,p |
| Energy | -887.15337424 a.u. |
| Gradient | 0.00001316 a.u. |
| Dipole Moment 2.21 Debye | |
| Point Group | C1 |
| Calculation Time | 01:42:59:2 |
Link to the files published to D-space for Compound 12 optimisation DOI:10042/23010
| Orbital number | MOPAC representation1 | MOPAC representation2 | DFT comparison | Comments |
|---|---|---|---|---|
| HOMO-2 |  |
 |
 |
|
| HOMO-1 |  |
 |
 |
|
| HOMO |  |
 |
 |
|
| LUMO |  |
 |
 |
|
| LUMO+1 |  |
 |
 |
|
| LUMO+2 |  |
 |
 |
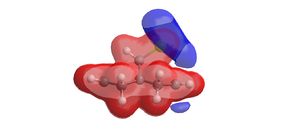 |
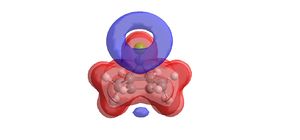
|
| Electrostatic potential surface of the bridged bicyclic compound | |

The blue region highlighted by the molecular electrostatic potential surface indicates an area of negative charge. It is clear that this region of negative charge is located above the double bond nearest the chlorine substitutent of the bridging carbon. This would strongly suggest that the reaction of dichlorocarbene is favoured to occur at this double bond on the upper face of the molecule.
Mechanism of Glycosidation

In this section is documented the results of a conformational analysis of various saccharide intermediate conformers in a glycosidation reaction. The two intermediates in question are the reactant and product of a single step in the three step reaction, shown right. The molecules were drawn in ChemBio3D and optimised first using the MM2 technique and then the PM6/MOPAC technique. The optimised structures along with the energy outputs from the optimisations are indicated in the tables below.
As can be seen from the second table the lowest energy conformation for the intermediate B was no .8. Whilst this wasn't formed the lowest energy conformation of compound A, the number 4 conformation, from which 8 is derived, in conformation 4 the carbonyl oxygen is closest to the reacting carbon centre and so the least amount of strain would likely be induced during the reaction, forming conformation 8. Thereby conformation 8 being the lowest energy. It can be seen that conformation 8 has significantly lower bond bending, torsion and 1,4-van der Waals repulsions which can be attributed to its precursor conformation already being in a similar conformation and so less molecular strain had to be invoked and weaker repulsive forces had to be overcome in order to form the five membered ring seen in compound B.
It should be noted that when performing the mopac energy minimisations, some of the substituents of the ring not involved in the reaction flipped from the equatorial position to axial positions. Despite tweeking the positions of the groups before the mopac minimsation some groups could not be prevented from changing position, and so the displayed structures are the lowest energy conformations achieved.
Simulation of Spectroscopic Data for a literature molecule
Preliminary Investigation of a Taxol Derivative

In preparation for using modelling software to predict spectroscopic data for a literature compound, an intermediate compound in the synthesis of taxol, see right, was investigated first.
Optimisation
After being drawn in ChemBio3D the energy of the intermediate, in the taxol synthesis, was minimised via the mm2 molecular mechanics technique before performing a subsequent optimisation using the 6-31G basis set and DFT-B3LYP method. This was performed in preparation for the NMR calculation.
Link to file published to D-space for the optimisation DOI:10042/23229
Predicted NMR Spectrum
Link to file published to d-space for Taxol synthesis intermediate NMR calculation DOI:10042/23227

Below are tabulated comparisons of the data for the predicted nmr and reported nmr shifts for both 1H NMR and 13. The literature values used for comparison are from here. [3]. For both sets of nmr shifts, the difference between the reported and predicted nmr shifts was less than 10% of the shift values, apart from a few cases (shift 1 1H nmr and shift 15 13C nmr) which would indicate close correlation between predicted and experimental shifts which provides strong evidence to the identity of compound 18 reported in the paper[3]. The outlying predicted shift, no 15, in the 13C NMR spectrum would likely suggest that the conformation about this single carbon atom differs from the compound reported. Given that the rest of the shifts correlate far better with the reported data, it might be the case that this error in conformation might have been introduced in drawing the molecule in the ChemBio3D program and could possibly be eradicated by further tweaking to find a lower energy conformation before performing the Gaussian based optimisation.
The NMR data obtained through the calculations performed would support the identity of compound 18 being the conformation reported.
| No. | 1H NMR Literature Shift Value/δppm | Comparable Theoretical Value/δppm | Difference in shift value | 1H NMR Literature Shift Value/δppm | Comparable Theoretical Value/δppm | Difference in Shift value |
|---|---|---|---|---|---|---|
| 1 | 1.03 | 0.82 | -0.21 | 22.2 | 22.5 | +0.3 |
| 2 | 1.07 | 1.00 | -0.07 | 25.4 | 23.5 | -1.9 |
| 3 | 1.1 | 1.22 | +0.12 | 25.6 | 24.8 | -0.8 |
| 4 | 1.35 | 1.43 | +0.08 | 30.00 | 30.1 | +0.1 |
| 5 | 1.58 | 1.66 | +0.08 | 30.8 | 30.5 | -0.3 |
| 6 | 1.95 | 1.98 | +0.03 | 35.5 | 34.4 | -0.1 |
| 7 | 2.53 | 2.48 | -0.05 | 36.8 | 35.4 | -1.4 |
| 8 | 2.85 | 2.76 | -0.09 | 38.7 | 39.2 | +0.5 |
| 9 | 5.21 | 5.50 | +0.31 | 40.8 | 41.1 | +0.3 |
| 10 | 43.3 | 45.8 | +2.5 | |||
| 11 | 45.5 | 46.9 | +1.4 | |||
| 12 | 50.9 | 50.0 | -0.9 | |||
| 13 | 51.3 | 53.5 | +2.2 | |||
| 14 | 60.5 | 61.2 | +0.7 | |||
| 15 | 74.6 | 87.4 | +12.8 | |||
| 16 | 120.9 | 124.7 | +3.8 | |||
| 17 | 148.7 | 152.5 | +3.8 | |||
| 18 | 211.5 | 211.4 | -0.1 |
Investigation of a stereospecific synthesis of 2-hydroxy-1-norbornanesulfonamide
This section of the wiki page details an investigation to the spectroscopic properties of a isomerically pure compound reported in literature. An ideal compound would be conformationally restricted, as the ability to adopt a range of conformations can make interpreting spectroscopic data more ambiguous as different conformers give rise to unique nmr shifts, optical rotations, absorption peaks etc. Thereby assignment of spectroscopic data to a single isomer becomes less accurate. Modelling programs such as Gaussian allow for prediction of the spectra that a molecule modelled in the program would give. The predicted spectra can be compared with published data to further investigate the identity of a prepared compound. Some differences are likely to arise between theoretical and experimental data but with the availability of high powered computing the has allowed for the complexity of the basis sets used to model compounds to be increased giving more accurate predictions, without having to endure extremely long reaction times. The compound being investigated is a 2-hydroxy-1-norbornanesulfonamide (2-hydroxy-1-NBSA) prepared by the stereoselective synthesis as reported by Martinez et al[4]. The ring and bridging component of the compound in question greatly restrict the possible conformations with this molecule making the modelling predictions a more valid technique and the data obtained more accurate. The variation in stereochemistry in the compound is at the 2 position, the hydroxyl carbon, where the hydroxyl group can be positioned in the axial or equatorial positions. The stereospecific control arises from the positioning of sterically hindering methyl groups on the ring or the bridge. However it is likely such a method of control would not give exclusively a single stereoisomer, as the steric hinderence that arises from the methyl groups would not be great as they are not bulky groups. So this investigation will attempt to identify whether the intended stereoisomer has been prepared exclusively or whether a mixture has been prepared.
Investigated 2-hydroxy-1-norbornanesulfonamide compound |
Optimisation
Link to file published to D-space for the optimisation DOI:10042/23151
The chosen molecule, shown right, was constructed in ChemBio3D and the energy minimised by the molecular mechanics method before creating a gaussian input file to be further optimised using the HPC.
NMR Calculations

The NMR calculations were performed using the 6-31G basis set Link to file published to D-space for the NMR calculation for the desired stereoisomer (reported) DOI:10042/23279
Link to file published to D-space for the NMR calculation for the alternate stereoisomer (unreported)DOI:10042/23226
1H NMR
The NMR calculation was performed using a modified input file created from the .log file from the prior optimisation. Below is tabulated 1H NMR data for both the predicted NMR spectrum and the published data. The protons responsible for the shifts were indicated on the spectrum produced. The data in the second column indicates the reported integral and multiplicity of the shift. The data in the fourth column indicates the predicted integral and the protons that give rise to the shift.
| No. | Literature Shift Value/δppm | Shift Data | Comparable Theoretical Value/δppm | Shift Data+responsible protons | Difference in shift value | Alternate stereoisomer predicted shifts/δppm |
|---|---|---|---|---|---|---|
| 1 | 1.1 | 1H, m | 1.12 | 1H, H39 | 0.04 | 1.14 |
| 2 | 1.17 | 3H, s | 1.19 | 2H, H36, H27 | 0.05 | 1.22 |
| 3 | 1.25 | 6H, d | 1.29 | 4H, H23, H26, H34, H37 | -0.04 | 1.30 |
| 4 | 1.39 | 3H, s | 1.47 | 3H, H28, H31, H38 | -0.08 | |
| 5 | 1.42 | 1H, m | 1.55 | 1H, H24 | -0.13 | 1.59 |
| 6 | 1.92 | 4H, m | 1.97 | 4H, H18, H21, H22, H35 | -0.05 | 1.97 |
| 7 | 2.36 | 1H, m | 2.27 | 2H, H19, H25 | 0.09 | 2.22 |
| 8 | n/a | n/a | 2.53 | 1H, H30 | n/a | 2.59 |
| 9 | 3.63 | 1H, m | 3.49 | 1H, H32 | 0.14 | 2.74 |
| 10 | 3.76 | 1H, d:septuplet | 3.93 | 1H, H33 | -0.17 | 3.21 |
| 11 | 4.09 | 1H, ddd | 4.01 | 1H, H40 | 0.08 | 3.97 |
| 12 | 4.25 | 1H, d | 4.47 | 1H, H20 | -0.22 | 5.04 |

There is close correlation between almost all of the pairs of shifts, with the notable exceptions being pairs, 8, 10 and 12, in terms of the shift position. With the remaining pairs being close to or less than 0.1 ppm apart. The relative integrals for the majority of the shifts have the same integrals with the exceptions being 2, 3 and 7. For the most part the multiplicities of the literature shifts agree but without a more detailed description of the literature splittings it is hard to confirm how closely the splittings match. A good example however of where they correlate very well is for shift no. 10, despite the difference in shift position, the assignment of H33 being responsible for a doublet of septuplets would as it can couple with the amine proton and the six methyl protons on the adjacent carbon atoms.
Despite some close matchings of literature shifts with predicted shifts there are discrepancies that would suggest that they can't be paired so well. Beginning with multiplicities, the literature shift reported at 1.17ppm is reported as a singlet. However the closest theoretical comparison, which is at 1.19ppm, on top of being a different integral arises, according to predictions from hydrogen atoms 36 and 27 which neighbour numerous other protons and so coupling would be expected. Upon inspecting the molecule there are no proton environments, of that degeneracy, that aren't within reasonable coupling distances from protons in a different environment. It is likely then that this singlet might be composed of several overlaid shifts of protons in similar environments. This might account for the differences in the integrals of the predicted and experiemental shifts. There is a difference in the integrals for shift pair 3, considering the integral and multiplicity this shift would be easily assigned to the methyl protons bound to carbon atoms 13 and 14. Given that the calculation has assigned this shift to the 4 protons, 23, 26, 3 and 37, this calls in to question the validity of the nuclei assignments given by the nmr calculation but also the applicability of the 6-31G d,p basis set for accurately determining the shifts of individual nuclei of this molecule and describing the couplings present. More accurate results, that would more decisively confirm the identity of the stereoisomer reported in the literature would be obtained from a 6-311G+2d,p basis set, as it would more accurately describe the electron density around the molecule.
The shift no 8, for which there is no literature comparison, is likely a proton which is close enough in shift to others that it has been overlaid with other shifts. However to difference in the theoretical shift position versus the experimentally observed shift for this single proton, most likely arising from the choice of basis set, the theoretical shift is not close enough to any others to be considered degenerate.
When compared with the shift positions of the alternate stereoisomer, with the hydroxyl group in the axial position, a much closer agreement with literature is observed for the equatorial hydroxyl group stereoisomer(reported stereoisomer) than the axial hydroxyl group stereoisomer. Differences in the correlation of multiplicities and couplings asside, this would suggest that the reported stereoisomer was the compound prepared.
13C NMR
| No. | Literature Shift Value/δppm | Comparable Theoretical Value/δppm | Difference in shift value/δppm | Alternate stereoisomer shift values/δppm | Difference in shift (alternate stereoisomer)/δppm | Shift Identity |
|---|---|---|---|---|---|---|
| 1 | 21.8 | 23.1 | 1.3 | 22.7 | +0.9 | C9 |
| 2 | 24.9 | 23.8 | +1.4 | 24.1 | -0.8 | C14 |
| 3 | 25 | 26.8 | +1.8 | 25.5 | 0.5 | C13 |
| 4 | 26.3 | 29.2 | +2.9 | 26.5 | 0.2 | C5 |
| 5 | 29.9 | 31.1 | -1.2 | 30.1 | 0.2 | C6 |
| 6 | 40.5 | 41.5 | -1 | 38.1 | -2.4 | C1 |
| 7 | 46.3 | 48.5 | -2.2 | 48.1 | 1.8 | C3 |
| 8 | 46.4 | 50.6 | -4.2 | 50.0 | 3.6 | C12 |
| 9 | 49.4 | 53.7 | -4.3 | 54.9 | 5.5 | C7 |
| 10 | 73.4 | 78.1 | -4.7 | 73.1 | -0.3 | C2 |
| 11 | 77.1 | 84 | -6.9 | 84.9 | 7.8 | C4 |

In contrast to the 1H NMR, the majority of data in the 13 shows a closer fit of the alternate stereoisomer predicted shifts to the reported literature values. In all but three cases the predicted shift for the alternate stereoisomer was closer to the liteature reported shifts than the predicted shifts for the equatorial-hydroxy stereoisomer. Whilst not a perfect correlation it does cast uncertainty over the true identity of the prepare compound.
In all cases the difference in shift between theoretical and reported values was less than %10 of the approximate shift value, an for many of the shifts even less, indicating that the predicted values show a good level of accuracy.
The split evidence provided by the NMR data shows that the exact identity of the prepared compound can't be confirmed, and shows the need for more complex basis sets to more accurately describe the electron distribution in the molecule and give more reliable shift values, couplings and assignments.
Frequency Analysis
Link to file published to D-space for the Frequency Analysis DOI:10042/23152
Vibrations
Below is a table of the most prominent vibrations observed in the predicted IR spectrum for the 2-hydroxyDue to the large number of vibrations, with significant intensity, that are predicted by the frequency analysis, a cut off point was determined so that a manageable number of vibrations could be investigated in depth. The cut off point was an intensity of 70, the N-H stretch vibration was included as it is a commonly reported absorption for amines and so was considered to be useful for The molecule has C1 symmetry and so all vibrations have the same symmetry label as only one is attainable, a.
| Frequency/cm-1 | Form of Vibration | Intensity |
|---|---|---|
| 571 | 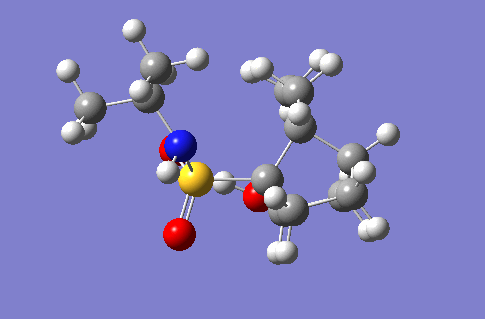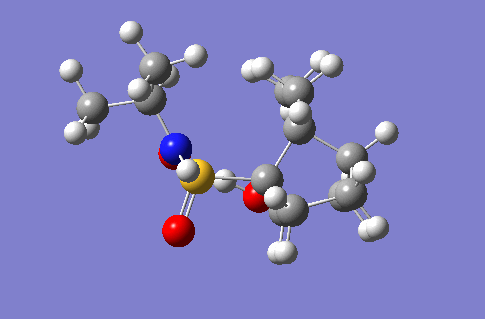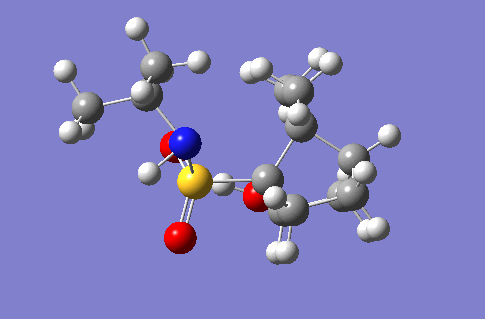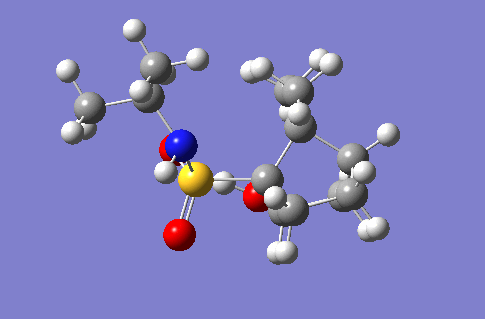 |
161.6 |
| 594 | 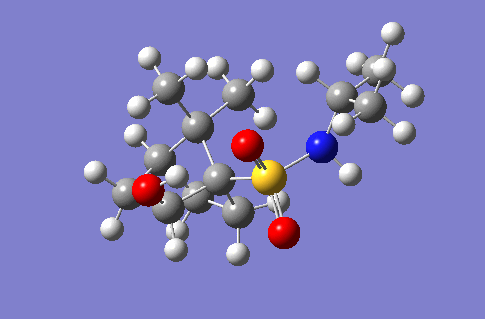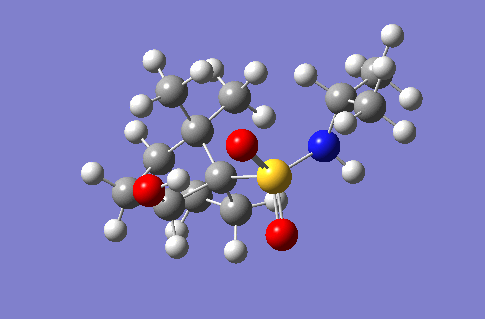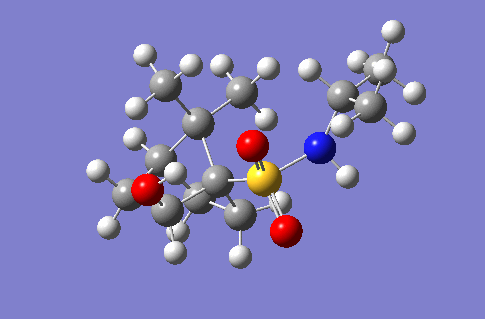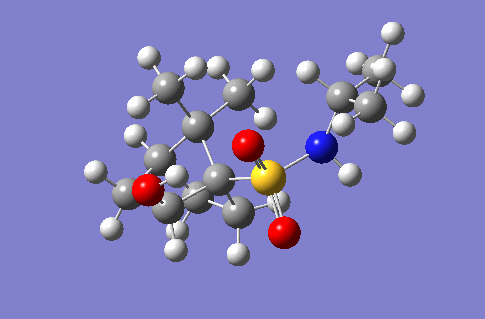 |
143.1 |
| 988 | 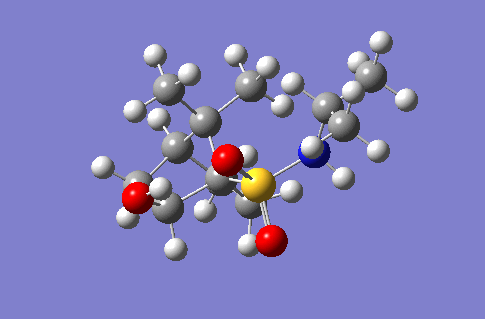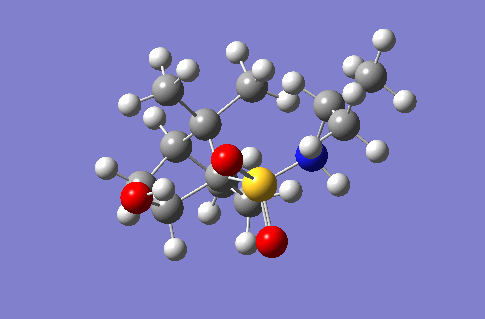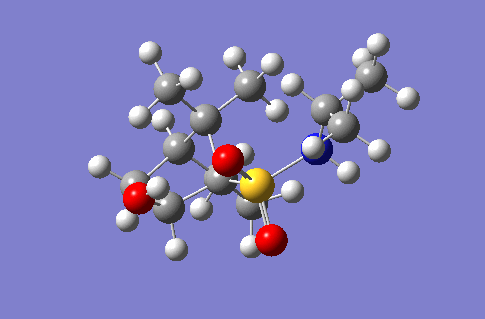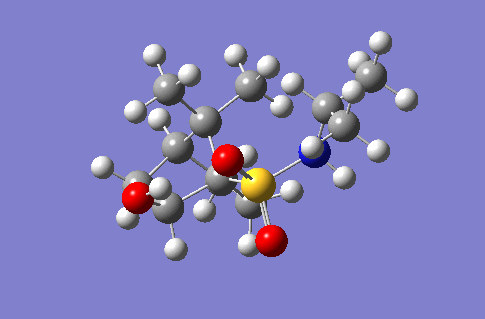 |
102.6 |
| 1086 | 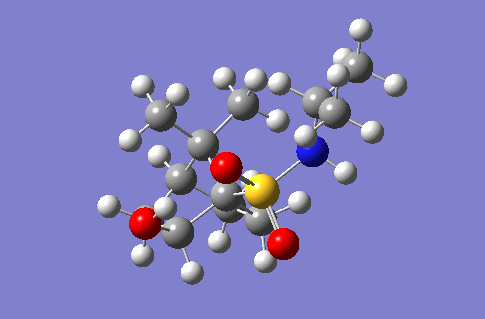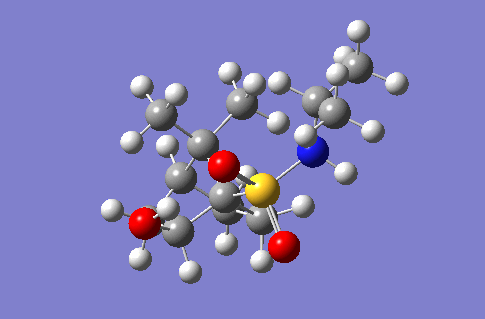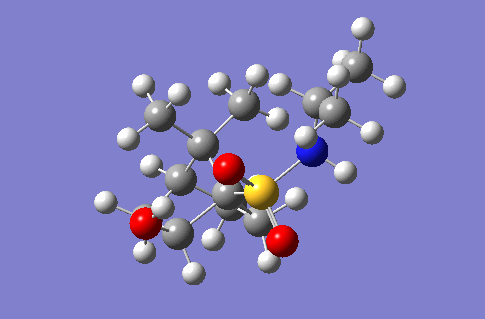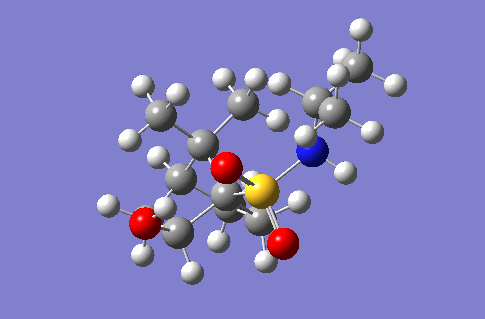 |
130.8 |
| 1121 |  |
106.1 |
| 1248 |  |
266.2 |
| 3540 |  |
56.7 |
| 3692 |  |
253.4 |
Despite many peaks appearing in the predicted IR spectrum, below, very few are actually reported in the literature for the compound. This might be that they weren't of significant enough intensity to warrant reporting, that they didn't serve any functional group identification purpose or that they weren't observed at all. Below are the tabulated predicted absorption frequencies and reported frequencies. The choice of comparison from the vast number of predicted vibrations was based on the intensity of the absorption, as well as its frequency. Whilst the predicted and reported frequencies are occur in the regions for which the functional groups giving rise to them are commonly found, there are large differences in the frequencies and so could not be readily used to confirm the identity of a particular stereoisomer.
| Reported Frequency/cm-1 | Comparable Predicted Frequency/cm-1 | Vibration Type |
|---|---|---|
| 1132 | 1121 | C-H bending |
| 1300 | 1248 | C-H bending, S=O stretch |
| 3333 | 3540 | N-H stretch |
| 3516 | 3692 | O-H stretch |
Predicted IR spectrum

References
- ↑ W.F. Maier & P. v. R. Schleyer. J. Am. Chem. Soc. 1981. 103. 1891-1900 [Evaluation and Prediction of the Stability of Bridgehead Olefins]
- ↑ A. B. McEwen & P. v. R. Schleyer. J. Am. Chem. Soc. 1986. 108 3951-3960[Hyperstable olefins:further calculation explorations and predictions]
- ↑ 3.0 3.1 L. A. Paquette, N. A. Pegg, D. Toops, G. D. Maynard, and R. D. Rogers. J. Am. Chem. Soc.. 1990. 112 277-283[of spectral data for taxol synthesis intermediate]
- ↑ A. G. Martı́nez, E. T. Vilar, F. M Jiménez, A. M. Á. Garcı́a.Tetrahedron-Asymmetr. 2004. 15. 293–298[short and convenient procedure for the stereoselective synthesis of 2-hydroxy-1-norbornanesulfonamides]























