Rep:Mod:benton1
Introduction
Computational chemistry is a powerful technique to analyse compounds. This is of particular use when these compounds are toxic, expensive or impractical to test manually using standard laboratory techniques. Recently, with advances in computing power and better models, computational chemistry has come to the fore as a tool to design better catalysts, and to analyse a wide range of ionic liquids which would be impractical to synthesis in their entirety, allowing their properties and reactivity to be analysed from the comfort of the researchers desk.
Part 1
A range of simple molecules are built, optimised and analysed using Guassview 09.
Borane Optimisation
Initial Optimisation
| BH3 | |||||
|---|---|---|---|---|---|
| |||||
| File Type | .log | ||||
| Log File | File:BH3 OPT2 log OB.LOG | ||||
| Calculation Type | FOPT | ||||
| Calculation Method | B3YLP | ||||
| Basis Set | 3-21G | ||||
| Final Energy | -26.46 a.u. | ||||
| Gradient | 0.00020672 a.u. | ||||
| Dipole Moment | 0.00 Debye | ||||
| Point Group | D3H | ||||
| CPU Time | 7.0 seconds | ||||
| B-H Bond Length | 1.19 a.u. | ||||
| H-B-H Bond Angle | 120.0 | ||||
Initially BH3 was drawn in with a trigonal planar geometry with 1.5Å B-H bond lengths. It was then optimised using a B3LYP method with a 3-21G basis set. Although this basis set has a very low accuracy, because of the small number of atoms and high level of symmetry (D3h) in borane, the basis set is sufficient.
The optimisation converged successfully as shown below.
Item Value Threshold Converged?
Maximum Force 0.000413 0.000450 YES
RMS Force 0.000271 0.000300 YES
Maximum Displacement 0.001610 0.001800 YES
RMS Displacement 0.001054 0.001200 YES
Predicted change in Energy=-1.071764D-06
Optimization completed.
-- Stationary point found.
Optimisation Procedure
Some Gumph about how it optiises by changing the schrodinger equation and iterates values.
6-31G(d,p) Optimisation
Using the geometry optimised above, the more accurate basis set 6-31G(d,p) was used to further refine the optimisation of BH3
| BH3 6-31G(d,p) | |||||
|---|---|---|---|---|---|
| |||||
| File Type | .log | ||||
| Log File | File:BH3 OPT 631 G OB.LOG | ||||
| Calculation Type | FOPT | ||||
| Calculation Method | RB3YLP | ||||
| Basis Set | 6-31G(d,p) | ||||
| Final Energy | -26.62 a.u. | ||||
| Gradient | 0.00000235 a.u. | ||||
| Dipole Moment | 0.00 Debye | ||||
| Point Group | D3H | ||||
| CPU Time | 8.0 seconds | ||||
| B-H Bond Length | 1.19 a.u. | ||||
| H-B-H Bond Angle | 120.0 | ||||
The optimisation converged successfully.
Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.000019 0.001800 YES
RMS Displacement 0.000012 0.001200 YES
Predicted change in Energy=-1.304276D-10
Optimization completed.
-- Stationary point found.
Comparison
These two results cannot be directly compared as they use different basis sets for the calculations, however both of the bond angles and lengths agree with literature values[1]
TlBr3
| TlBr3 | |||||
|---|---|---|---|---|---|
| |||||
| File Type | .log | ||||
| Log File | File:Log 69185.log | ||||
| D-Space | DOI:10042/22681 | ||||
| Calculation Type | FOPT | ||||
| Calculation Method | RB3YLP | ||||
| Basis Set | LANL2DZ | ||||
| Final Energy | -91.22 a.u. | ||||
| Gradient | 0.00000090 a.u. | ||||
| Dipole Moment | 0.00 Debye | ||||
| Point Group | D3H | ||||
| CPU Time | 28.3 seconds | ||||
| Tl-Br Bond Length | 2.65 a.u. | ||||
| Br-Tl-Br Bond Angle | 120.0 | ||||
TlBr3 was constructed using Gaussview, and its symmetry restricted to D3h (with a tolerance of 0.0001). A medium level basis set was used for the optimisation, that uses pseudo potentials with heavier elements. With over 186 electrons, TlBr3 displays relativistic effects which the standard Schrodinger equation doesn't recover. The idea is that there is little (to none) overlap between the core electrons and the valence electrons. These core electrons are not involved in bonding and the normal Coulombic term that would represent them in the Schrodinger equation is modified. This has the benefit of drastically reducing the number of electrons involved in calculations; hence a smaller basis set is needed.
The molecule successfully converged.
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000014 0.001200 YES
Predicted change in Energy=-6.084002D-11
Optimization completed.
-- Stationary point found.
BBr3
| BBr3 | ||
|---|---|---|
 | ||
| File Type | .log | |
| Log File | File:Log 69204.log | |
| D-Space | DOI:10042/22680 | |
| Calculation Type | FOPT | |
| Calculation Method | RB3YLP | |
| Basis Set | Gen | |
| Final Energy | -64.44 a.u. | |
| Gradient | 0.00000382 a.u. | |
| Dipole Moment | 0.00 Debye | |
| Point Group | D3H | |
| CPU Time | 16.4 seconds | |
| Tl-Br Bond Length | 1.93 a.u. | |
| Br-Tl-Br Bond Angle | 120.0 | |
BBr3, as it combines light and heavier elements needs a combination of pseudo potentials and basis sets. The BH3 optimised using the 6-31G(d,p)basis set was used as a starting point to construct the BBr3 molecule.
The molecule successfully converged.
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000036 0.001800 YES
RMS Displacement 0.000023 0.001200 YES
Predicted change in Energy=-4.026780D-10
Optimization completed.
-- Stationary point found.
Comparing and Contrasting BH3 BBr3 and TlBr3
| Molecule | Bond Distance (a.u.) |
|---|---|
| BH3 | 1.19 |
| BBr3 | 1.93 |
| TlBr3 | 2.65 |
As the molecules get heavier and bigger, the bonds get longer. This pattern is observed throughout chemistry and is because further down a group the atoms are, the more higher their nuclear charge and the higher the number of electrons. these electrons, due to several factors including shielding, electrostatic repulsion, and the Pauli exclusion principle. H and Br are similar in that they both require one electron to 'fill' their molecular orbitals and so they have a full valence orbitals and are at their msot stable. How they differ however is that the hydrogen bonds are purely s in character, due to the nature of the hydrogen atom and only having one electron. The bonds with the Br atom, by contrast will have some p character due to the position of Br in the periodic table; it requires a p electron. The valence atomic orbitals in Br are a lot more diffuse than the s orbital in H due to the larger size of the atom. Br is also a lot heavier than H, and this will, coupled with the large increase in size will drastically effect the nature of bonds made. Within Guassview, a bond is determined by physical, through space distance. While for most molecules this gross approximation is sufficient, in some circumstances it does not represent the tru nature of the interactions between the molecules. A chemical bond is a strong electrostatic attraction between two or more molecules, with a high electron density between the two nuclei which binds them together.
Frequency Analysis
With knowledge gleaned from a frequency analysis, an IR spectra can be computed, predicting what stretches will occur and at what energies.
BH3
The previously optimised (using the 6-31G(d,p) basis set) BH3 molecule was analysed using Guassian.
| BH3 | |||||
|---|---|---|---|---|---|
| |||||
| File Type | .log | ||||
| Log File | File:OHB BH3 FREQ.LOG | ||||
| Calculation Type | FREQ | ||||
| Calculation Method | RB3YLP | ||||
| Basis Set | 6-31G(d,p) | ||||
| Final Energy | -26.62 a.u. | ||||
| Gradient | 0.00000237 a.u. | ||||
| Dipole Moment | 0.00 Debye | ||||
| Point Group | D3H | ||||
| CPU Time | 7.0 seconds | ||||
| B-H Bond Length | 1.19 a.u. | ||||
| H-B-H Bond Angle | 120.0 | ||||
Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000002 0.000300 YES
Maximum Displacement 0.000019 0.001800 YES
RMS Displacement 0.000009 0.001200 YES
Predicted change in Energy=-1.323374D-10
Optimization completed.
-- Stationary point found.
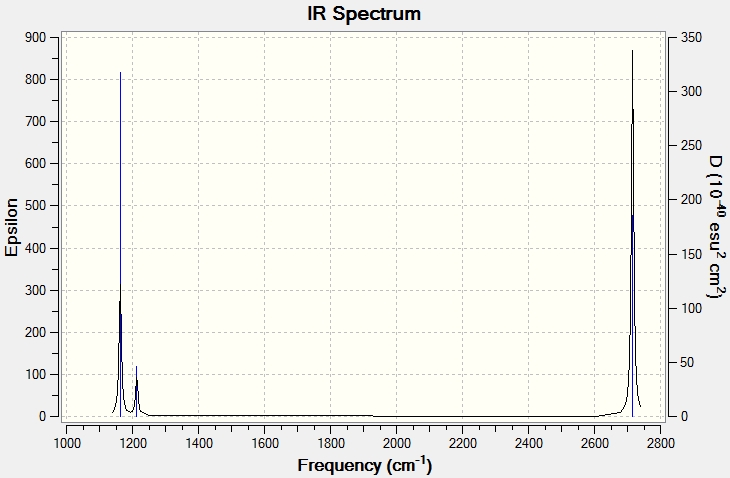
Low frequencies --- -0.9033 -0.7343 -0.0054 6.7375 12.2491 12.2824 Low frequencies --- 1163.0003 1213.1853 1213.1880
| Number | Form of Vibration | Description | Frequency | Intensity | Symmetry D3H | |||
|---|---|---|---|---|---|---|---|---|
| 1 |
|
Wag - All Hydrogens remain in plane and move together perpendicular to it, B moves in opposite direction | 1163.00 | 92.54 | A2" | |||
| 2 |
|
Two hydrogens scissoring, other H and Boron moving away | 1213.19 | 14.06 | E' | |||
| 3 |
|
Two hydrogens scissoring, third hydrogen moving laterally, boron counteracting motion. All in the same plane. | 1213.19 | 14.06 | E' | |||
| 4 |
|
All three hydrogens symmetrically stretching | 2582.26 | 0.00 | A1' | |||
| 5 |
|
Two hydrogens asymetrically stretching | 2715.43 | 126.33 | E' | |||
| 6 |
|
Two hydrogens symmetrically stretching with each other, asymetrically with the third | 2715.43 | 126.33 | E' |
While there are six vibrational modes, (see table, 'above') there are only three vibrations shown on the computed IR spectrum. This is for two reasons. Firstly, one of the vibrations has an intensity of 0, so does not show up on the spectrum as it is the same as the baseline. Secondly, there are two sets of two vibrations that occur at the same frequency and so the four vibrations, (modes 1 & 2 and modes 3 & 4) only show up as two vibrations.
TlBr3
The previously optimised TlBr3 molecule was analysed.
| TlBr3 | |||||
|---|---|---|---|---|---|
| |||||
| File Type | .log | ||||
| Log File | File:Ohb tlbr3 freq.log | ||||
| D-Space | DOI:10042/22683 | ||||
| Calculation Type | Freq | ||||
| Calculation Method | RB3YLP | ||||
| Basis Set | LANL2DZ | ||||
| Final Energy | -91.22 a.u. | ||||
| Gradient | 0.00000088 a.u. | ||||
| Dipole Moment | 0.00 Debye | ||||
| Point Group | D3H | ||||
| CPU Time | 28.5 seconds | ||||
| Tl-Br Bond Length | 2.65 a.u. | ||||
| Br-Tl-Br Bond Angle | 120.0 | ||||
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000011 0.001200 YES
Predicted change in Energy=-5.660901D-11
Optimization completed.
-- Stationary point found.
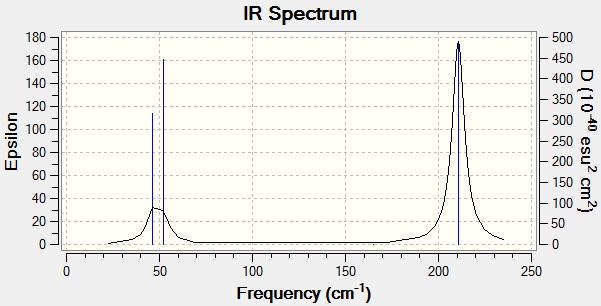
Low frequencies --- -3.4213 -0.0026 -0.0004 0.0015 3.9367 3.9367 Low frequencies --- 46.4289 46.4292 52.1449
| Number | Form of Vibration | Description | Frequency (cm-1) | Intensity | Symmetry D3H | |||
|---|---|---|---|---|---|---|---|---|
| 1 |
|
Two Br scissoring, other Br and Boron moving away | 46.43 | 3.69 | E' | |||
| 2 |
|
Rocking in plane. | 46.43 | 3.69 | E' | |||
| 3 |
|
Wag - All Br remain in plane and move together perpendicular to it, Tl moves in opposite direction | 52.14 | 3.69 | A2" | |||
| 4 |
|
All three Br symmetrically stretching | 165.27 | 0.00 | A1' | |||
| 5 |
|
Two Br asymetrically stretching | 210.69 | 126.33 | E' | |||
| 6 |
|
Two Br symmetrically stretching with each other, asymetrically with the third | 210.69 | 126.33 | E' |
While there are six vibrational modes, (see table) there are only three vibrations shown on the computed IR spectrum. One of the vibrations has an intensity of 0, so does not show up on the spectrum as it is the same as the baseline. It has an intensity of 0 as it is a symmetrical stretch and does not change the dipole, it is therefore IR inactive.
Secondly, there are two sets of two vibrations that are degenerate, so the four vibrations, (modes 1 & 2 and modes 3 & 4) only show up as two vibrations.
Comparison of Frequencies and Analysis
| Symmetry | TlBr3 Frequency (cm-1) | BH3 Frequency (cm-1) | |
|---|---|---|---|
| E' | 46.43 | 1213.19 | |
| E' | 46.43 | 1213.19 | |
| A2" | 52.14 | 1163.00 | |
| A1' | 165.27 | 2582.26 | |
| E' | 210.69 | 2715.43 | |
| E' | 210.69 | 2715.43 |
There is a large difference in the frequencies between TlBr3 and BH3, over an order of magnitude difference. This can be explained by the effect of the reduced mass on the effect of vibrational frequencies. Using the approximation of Hooke's law to model the vibrational modes, where
Thus the magnitude of the vibrational modes is inversely proportional to the square root of .
,
and so for BH3 is 0.92, whereas for TlBr3 it is 57, a much higher value. Another reason is that the B-H bond is a lot stronger than the Tl-Br bond. In BH3, the orbitals overlap significantly, producing a stronger bond, whereas in the TlBr3 analogue, because of the larger, more diffuse orbitals, the overlap is poorer, leading to a weaker bond. Both spectra display three peaks, two of these are the result of the summation of two degenerate vibrational modes. In both spectra the A2' and E' modes lie close together and at a higher energy, the A1' and E' modes lie close together. the large E gap between the two groups of modes, which is present in both molecules is due to the different amount of energy required to bend or stretch a molecule. Bending requires significantly lower energy to occur and as such all of the bending modes occur at lower frequencies. The ordering of the modes is however switched between the two molecules, as highlighted by the diagram. It is switched due to the different masses of the elements involved; as Br is a lot more massive than H, it requires more energy to perform the out of plane bending required of the A2' mode. The same logic can be applied as to why the A1' mode is lower than the E' modes for the BH3 molecule; due to the relative lightness of the hydrogen atom it requires very little energy to move it out of the plane of the atom, and hence happens at a lower frequency.
The same method and basis set must be used for both the optimisation and frequency calculations because the method used and quality of the basis set used directly effect the molecules final energy. If different methods and basis sets are used it is like comparing apples and oranges; different methods and quality of starting assumptions have been used and so will generate a different result; the end products are not comparable.
The "low frequencies" in the frequency calculation represent the molecules centre of mass. If a non linear molecule has 3N-6 normal vibrational modes
[2]
the low frequencies represent the -6 of the number of modes.

Mo Analysis
BH3
The previously optimised (#6-31G(d,p) Optimisation) BH3 molecule had it's MOs calculated.
| BH3 | ||||
|---|---|---|---|---|
| ||||
| File Type | .fch | |||
| Log File | File:Bh3 opt 631 g OB.chk | |||
| Calculation Type | SP | |||
| Calculation Method | RB3YLP | |||
| Basis Set | 6-31G(D,P) | |||
| Final Energy | -26.62 a.u. | |||
| Gradient | 0.00000000 a.u. | |||
| Dipole Moment | 0.00 Debye | |||
| Point Group | ||||
| CPU Time | 8.0 seconds | |||
| B-H Bond Length | 1.19 a.u. | |||
| H-B-H Bond Angle | 120.0 | |||
The calculated and qualitated MO diagrams show very similar if not exactly the same result. The Calculated MO's are more spread out, highlighting the more diffuse nature, however the differences are small; this shows the power of qualitative MO theory and how well it relates to high power comptational methods.
NBO analysis
NH3 was optimised using the 6-31G(d,p) basis set. It is appropriate to skip the initial optimisation that was performed for BH3 as NH3 is a smaller molecule. Frequency analysis was also carried out to ensure a minimum was obtained.
| NH3 | ||||||
| Molecule |
| |||||
|---|---|---|---|---|---|---|
| File Type | .log | .log | .log | |||
| Log File | File:OhbNH3OPT.LOG | File:OHBNH3FREQ.LOG | File:OHBNH3NBO.LOG | |||
| Calculation Type | FOPT | FREQ | SP | |||
| Calculation Method | RB3YLP | RB3YLP | RB3YLP | |||
| Basis Set | 6-31G(d,p) | 6-31G(d,p) | 6-31G(d,p) | |||
| Final Energy | -56.56 a.u. | -56.56 a.u. | -56.56 a.u. | |||
| Gradient | 0.00000289 a.u. | 0.00000281 a.u. | - | |||
| Dipole Moment | 1.8464 Debye | 1.8464 Debye | 1.8464 Debye | |||
| Point Group | C3V | C3 | C3V | |||
| CPU Time | 13.0 seconds | 8.0 seconds | 3.0 seconds | |||
The outputs are summarised below
Optimisation
Item Value Threshold Converged? Maximum Force 0.000005 0.000450 YES RMS Force 0.000003 0.000300 YES Maximum Displacement 0.000010 0.001800 YES RMS Displacement 0.000007 0.001200 YES Predicted change in Energy=-7.830780D-11 Optimization completed.
Frequency analysis
Low frequencies --- -11.6313 -11.5960 -0.0028 0.0243 0.1402 25.5608 Low frequencies --- 1089.6620 1694.1733 1694.1736
Item Value Threshold Converged? Maximum Force 0.000005 0.000450 YES RMS Force 0.000003 0.000300 YES Maximum Displacement 0.000011 0.001800 YES RMS Displacement 0.000006 0.001200 YES Predicted change in Energy=-8.408692D-11
NBO Analysis


(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.99909) BD ( 1) N 1 - H 2
( 68.83%) 0.8297* N 1 s( 24.87%)p 3.02( 75.05%)d 0.00( 0.09%)
0.0001 0.4986 0.0059 0.0000 0.0000
0.0000 0.8155 0.0277 -0.2909 0.0052
0.0000 0.0000 -0.0281 -0.0087 0.0013
( 31.17%) 0.5583* H 2 s( 99.91%)p 0.00( 0.09%)
0.9996 0.0000 0.0000 -0.0289 0.0072
2. (1.99909) BD ( 1) N 1 - H 3
( 68.83%) 0.8297* N 1 s( 24.87%)p 3.02( 75.05%)d 0.00( 0.09%)
0.0001 0.4986 0.0059 0.0000 -0.7062
-0.0240 -0.4077 -0.0138 -0.2909 0.0052
0.0076 0.0243 0.0140 0.0044 0.0013
( 31.17%) 0.5583* H 3 s( 99.91%)p 0.00( 0.09%)
0.9996 0.0000 0.0250 0.0145 0.0072
3. (1.99909) BD ( 1) N 1 - H 4
( 68.83%) 0.8297* N 1 s( 24.87%)p 3.02( 75.05%)d 0.00( 0.09%)
0.0001 0.4986 0.0059 0.0000 0.7062
0.0240 -0.4077 -0.0138 -0.2909 0.0052
-0.0076 -0.0243 0.0140 0.0044 0.0013
( 31.17%) 0.5583* H 4 s( 99.91%)p 0.00( 0.09%)
0.9996 0.0000 -0.0250 0.0145 0.0072
4. (1.99982) CR ( 1) N 1 s(100.00%)
1.0000 -0.0002 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
5. (1.99721) LP ( 1) N 1 s( 25.38%)p 2.94( 74.53%)d 0.00( 0.10%)
0.0001 0.5036 -0.0120 0.0000 0.0000
0.0000 0.0000 0.0000 0.8618 -0.0505
0.0000 0.0000 0.0000 0.0000 -0.0310
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (H3N)
1. BD ( 1) N 1 - H 2 1.99909 -0.60417
2. BD ( 1) N 1 - H 3 1.99909 -0.60417
3. BD ( 1) N 1 - H 4 1.99909 -0.60417
4. CR ( 1) N 1 1.99982 -14.16767
5. LP ( 1) N 1 1.99721 -0.31755 16(v),20(v),24(v),17(v)
21(v),25(v)
The natural bond order analysis shows where the electrons are situated and how the bonding occurs within the molecule. The information above shows that the three N-H bonds are equal in energy, and the lone pair has a high energy and all are occupied by two electrons; ie the bonds are 2c-2e bonds. It also hiughlight that the nitrogen bonds are sp3 hybridised as each one has a 24.87% S character and a 75.05% p character.
NH3BH3
A molecule of NH3BH3 was optimised to the 6-31G(d,p) level using the B3YLP method.
| NH3 | ||||||
| Molecule |
| |||||
|---|---|---|---|---|---|---|
| File Type | .log | .log | .log | |||
| Log File | File:OHBNB1.LOG | File:OHBNB2.LOG | File:OHBNB3.LOG | |||
| Calculation Type | FOPT | FOPT | FREQ | |||
| Calculation Method | RB3YLP | RB3YLP | RB3YLP | |||
| Basis Set | 3-21G | 6-31G(d,p) | 6-31G(d,p) | |||
| Final Energy | -82.7666 a.u. | -83.2247 a.u. | -83.2247 a.u. | |||
| Gradient | 0.00003006 a.u. | 0.00005659 a.u. | 0.00005643 a.u. | |||
| Dipole Moment | 5.8431 Debye | 5.5623 Debye | 5.5623 Debye | |||
| Point Group | C1 | C1 | C1 | |||
| CPU Time | 29.0 seconds | 30.0 seconds | 38.0 seconds | |||
3-21G Optimisation
Item Value Threshold Converged?
Maximum Force 0.000094 0.000450 YES
RMS Force 0.000030 0.000300 YES
Maximum Displacement 0.000419 0.001800 YES
RMS Displacement 0.000178 0.001200 YES
Predicted change in Energy=-5.742842D-08
Optimization completed.
-- Stationary point found.
6-31G(d,p) Optimisation
Item Value Threshold Converged?
Maximum Force 0.000133 0.000450 YES
RMS Force 0.000037 0.000300 YES
Maximum Displacement 0.001280 0.001800 YES
RMS Displacement 0.000567 0.001200 YES
Predicted change in Energy=-1.199663D-07
Optimization completed.
-- Stationary point found.
Frequency
Low frequencies --- -0.0013 -0.0010 0.0010 9.3030 12.2326 19.6429 Low frequencies --- 263.2969 631.3087 637.9206
Item Value Threshold Converged?
Maximum Force 0.000254 0.000450 YES
RMS Force 0.000056 0.000300 YES
Maximum Displacement 0.001417 0.001800 YES
RMS Displacement 0.000712 0.001200 YES
Predicted change in Energy=-2.159749D-07
Optimization completed.
-- Stationary point found.
Comparison of reaction energies
| Species | Energy (a.u.) |
|---|---|
| NH3 | -56.5578 |
| BH3 | -26.6153 |
| BH3NH3 | -83.2247 a.u. |
The difference in energy between the two reactants and the Lewis acid-base adduct is 135.4758kJ/mol
Part 2 - Ionic Liquids
Ionic liquids are molten ionic salts which are liquids at room temperature. They have some quite novel properties, and are being researched for use in liquid flow supercapacitors [3]
Comparison of cations
[N(CH3)4 ]+
| [N(CH3)4 ]+ | |||||||
| Molecule |
| ||||||
|---|---|---|---|---|---|---|---|
| File Type | .log | .log | .log | .fch | |||
| Log File | File:OhbN1 321log 69500.log | File:OhbN2 631log 69503.log | File:OhbN3 FREQlog 69513.log | File:OhbN4 MOlog 69525.log | |||
| D-Space | DOI:10042/22661 | DOI:10042/22662 | DOI:10042/22663 | DOI:10042/22664 | |||
| Calculation Type | FOPT | FOPT | FREQ | SP | |||
| Calculation Method | RB3YLP | RB3YLP | RB3YLP | RB3YLP | |||
| Basis Set | 3-21 | 6-31G(d,p) | 6-31G(d,p) | 6-31G(d,p) | |||
| Final Energy | -213.0164 a.u. | -214.1813 a.u. | -214.1813 a.u. | -214.1813 a.u. | |||
| Gradient | 0.00000763 a.u. | 0.00006797 a.u. | 0.00006800 a.u. | - a.u. | |||
| Dipole Moment | 22.2224 Debye | 22.2224 Debye | 22.2224 Debye | 0.0005 Debye | |||
| Point Group | C1 | C1 | C1 | C1 | |||
| CPU Time | 1 minute 12.8 seconds | 2 minutes 28.8 seconds | 6 minutes 36.5 seconds | 58.3 seconds | |||
The outputs are summarised below [N(CH3)4 ]+
3-21 Basis Set Optimisation
Item Value Threshold Converged?
Maximum Force 0.000031 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000641 0.001800 YES
RMS Displacement 0.000206 0.001200 YES
Predicted change in Energy=-7.454775D-09
Optimization completed.
-- Stationary point found.
6-31G(d,p)Basis Set Optimisation
Item Value Threshold Converged?
Maximum Force 0.000129 0.000450 YES
RMS Force 0.000049 0.000300 YES
Maximum Displacement 0.000809 0.001800 YES
RMS Displacement 0.000302 0.001200 YES
Predicted change in Energy=-2.842702D-07
Optimization completed.
-- Stationary point found.
6-31G(d,p)Basis Set Frequency Calculation
Full mass-weighted force constant matrix: Low frequencies --- -19.2061 -12.7564 -8.2164 -0.0007 -0.0001 0.0007 Low frequencies --- 174.6301 275.6794 282.5852
Item Value Threshold Converged?
Maximum Force 0.000132 0.000450 YES
RMS Force 0.000068 0.000300 YES
Maximum Displacement 0.000603 0.001800 YES
RMS Displacement 0.000305 0.001200 YES
Predicted change in Energy=-2.759911D-07
Optimization completed.
-- Stationary point found.
[P(CH3)4 ]+
| [P(CH3)4 ]+ | |||||||
| Molecule |
| ||||||
|---|---|---|---|---|---|---|---|
| File Type | .log | .log | .log | .log | |||
| Log File | File:OHBP1 321log 69501.log | File:OHBP2 631log 69504.log | File:OHBP3freq2ultrafine log 69624.log | File:OHBP4 MOlog 69527.log | |||
| D-Space | DOI:10042/22667 | DOI:10042/22668 | DOI:10042/22669 | DOI:10042/22670 | |||
| Calculation Type | FOPT | FOPT | FREQ | SP | |||
| Calculation Method | RB3YLP | RB3YLP | RB3YLP | RB3YLP | |||
| Basis Set | 3-21 | 6-31G(d,p) | 6-31G(d,p) | 6-31G(d,p) | |||
| Final Energy | -498.2055 a.u. | -500.8270 a.u. | -500.8270 a.u. | -500.8270 a.u. | |||
| Gradient | 0.00002920 a.u. | 0.00001371 a.u. | 0.00000071 a.u. | - a.u. | |||
| Dipole Moment | 22.2188 Debye | 22.2177 Debye | 22.2223 Debye | 0.0022 Debye | |||
| Point Group | C1 | C1 | C1 | C1 | |||
| CPU Time | 1 minute 51.2 seconds | 3 minutes 20.0 seconds | 18 minutes 24.9 seconds | 56.0 seconds | |||
The outputs are summarised below [N(CH3)4 ]+
3-21 Basis Set Optimisation
Item Value Threshold Converged?
Maximum Force 0.000114 0.000450 YES
RMS Force 0.000027 0.000300 YES
Maximum Displacement 0.000685 0.001800 YES
RMS Displacement 0.000236 0.001200 YES
Predicted change in Energy=-1.277134D-07
Optimization completed.
-- Stationary point found.
6-31G(d,p)Basis Set Optimisation
Item Value Threshold Converged?
Maximum Force 0.000051 0.000450 YES
RMS Force 0.000016 0.000300 YES
Maximum Displacement 0.000987 0.001800 YES
RMS Displacement 0.000310 0.001200 YES
Predicted change in Energy=-5.307280D-08
Optimization completed.
-- Stationary point found.
6-31G(d,p)Basis Set Frequency Calculation
Full mass-weighted force constant matrix: Low frequencies --- -2.6330 0.0021 0.0028 0.0030 5.1381 7.5743 Low frequencies --- 156.4446 192.0398 192.2775
Item Value Threshold Converged? Maximum Force 0.000001 0.000015 YES RMS Force 0.000000 0.000010 YES Maximum Displacement 0.000098 0.000060 NO RMS Displacement 0.000032 0.000040 YES Predicted change in Energy=-8.257063D-11
Although the above highlights that this calculation did not converge to the tight tolerences set for the maximum displacement, this calculation continued to be used. The value, while larger than the threshold set, is still over an order of magnitude smaller that the normal threshold value. The low frequencies are also very close to 0,which is also indicative that the calculation has run correctly, and so the result can be relied on to the degree of accuracy required.
[S(CH3)3]+
| [S(CH3)3 ]+ | |||||||
| Molecule |
| ||||||
|---|---|---|---|---|---|---|---|
| File Type | .log | .log | .log | .log | |||
| Log File | File:OhbS1log 69541.log | File:OHBS2log 69556.log | File:S3FREQlog 69561.log | File:S4MOlog 69564.log | |||
| D-Space | DOI:10042/22678 | DOI:10042/22679 | DOI:10042/22844 | DOI:10042/22843 | |||
| Calculation Type | FOPT | FOPT | FREQ | SP | |||
| Calculation Method | RB3YLP | RB3YLP | RB3YLP | RB3YLP | |||
| Basis Set | 3-21 | 6-31G(d,p) | 6-31G(d,p) | 6-31G(d,p) | |||
| Final Energy | -515.0523 a.u. | -517.6833 a.u. | -517.6833 a.u. | XXX a.u. | |||
| Gradient | 0.00004889 a.u. | 0.00002584 a.u. | 0.00002542 a.u. | 0.00000XXX a.u. | |||
| Dipole Moment | 1.2886 Debye | 0.9652 Debye | 0.9652 Debye | xxx Debye | |||
| CPU Time | 2 minuteS 0.0 seconds | 3 minutes 59.4 seconds | 3 minutes 37.9 seconds | XX seconds | |||
3-21 Basis Set Optimisation
Item Value Threshold Converged?
Maximum Force 0.000159 0.000450 YES
RMS Force 0.000044 0.000300 YES
Maximum Displacement 0.001697 0.001800 YES
RMS Displacement 0.000572 0.001200 YES
Predicted change in Energy=-2.884895D-07
Optimization completed.
-- Stationary point found.
6-31G(d,p)Basis Set Optimisation
Item Value Threshold Converged?
Maximum Force 0.000053 0.000450 YES
RMS Force 0.000023 0.000300 YES
Maximum Displacement 0.001368 0.001800 YES
RMS Displacement 0.000420 0.001200 YES
Predicted change in Energy=-6.993418D-08
Optimization completed.
-- Stationary point found.
6-31G(d,p)Basis Set Frequency Calculation
Full mass-weighted force constant matrix: Low frequencies --- -23.9260 -14.4954 -0.0038 -0.0031 -0.0025 10.3677 Low frequencies --- 161.5670 195.2390 207.2525
The values for the frequencies are a little higher than is ideal. However, because the basis set is too small for sulphur to be calculated accurately, the reasonably large values of -23.9260 and -14.4954 are acceptable. These frequencies represent the movement of the centre of mass; translations and rotations.
Comparison of NBOs
The CH bonds in all the molecules remain almost the same, however the bond between the C-E (where E= N,P,S)changes depending on the nature of E. The nitrogen contributes more to the C-E bond than the phosphorous does as the atomic orbitals on N are less diffuse and overlap better with the carbon. Both N and P are completely sp3 hybridised, as could be rationalised from the tetrahedral shapes. The S compound is a distorted tetrahedral, with a lone pair occupying one of the tetrahedral sites.
| Molecule | Atom | Charge |
|---|---|---|
| [N(CH3)4 ]+ | N | -0.295 |
| C | -0.483 | |
| H | 0.269 | |
| [P(CH3)4 ]+ | P | 0.726 |
| C | -0.511 | |
| H | 0.193 | |
| [S(CH3)3 ]+ | S | 0.917 |
| C | -0.845 | |
| H | 0.279-0.297 |
As is highlighted above, the distribution of charge changes depending on the nature of E. In the Phosphorous analogue, the positive charge is mostly centered on the phosphorous atom, whereas the in the N molecule, the positive charge is distributed around the methyl-hydrogens around the edge of the molecule, with a negative core. As nitrogen is more electronegative than phosphorous the atom holds onto its charge better; the phosphorous atom more readily gives up its electrons to the rest of the electron deficient structure. This also links to the population analyses above; because the N atom is more electronegative it is more able to donate more electrons into the 2c-2e C-E bond, whereas because phosphorous int he cation is more electron deficient than in the uncharged species, it is able to contribute less to the C-E bond. The population analysis shows that the with the sulphur compound, most of the positive charge is on the sulphur atom. Despite being of similar electronegativity to carbon, the sulphur atom has a lone pair and so is more easily able to lose an electron. In comparison to the simplified picture of NR4+, where the positive charge resides purely on the nitrogen. This picture arises from a lewis structure of the bonding, that the lone pair of the acts as a nucleophile and bonds to a methyl cation, forming a tetravalent N+ compound. This picture is not correct however, as shown by the NBO analysis. The positive charge is actually distributed equally among the hydrogens on the exterior of the molecule, leaving a negative, neucleophilic centre. This can be rationalised that as nitrogen very electronegative, it draws electrons towards it, leaving the exterior, which is further away, more positive.
Influence of Functional Groups
[N(CH3)3CH2CN]+
| [N(CH3)3CH2CN]+ | |||||||
| Molecule |
| ||||||
|---|---|---|---|---|---|---|---|
| File Type | .log | .log | .log | .log | |||
| Log File | File:OHBNCN1 321log 69612.log | File:OHBNCN2 631log 69620.log | File:OHBNCN3 FREQ2log 69647.log | File:OHBNCN4 MOlog 69675.log | |||
| D-Space | DOI:10042/22686 | DOI:10042/22687 | DOI:10042/22688 | DOI:10042/22689 | |||
| Calculation Type | FOPT | FOPT | FREQ | SP | |||
| Calculation Method | RB3YLP | RB3YLP | RB3YLP | RB3YLP | |||
| Basis Set | 3-21 | 6-31G(d,p) | 6-31G(d,p) | 6-31G(d,p) | |||
| Final Energy | -304.7232 a.u. | -306.3938 a.u. | -306.3938 a.u. | -306.3938 a.u. | |||
| Gradient | 0.00002973 a.u. | 0.00001171 a.u. | 0.00000027 a.u. | - a.u. | |||
| Dipole Moment | 24.5324 Debye | 24.6488 Debye | 24.6491 Debye | 24.6488 Debye | |||
| Point Group | C1 | C1 | C1 | C1 | |||
| CPU Time | 3 minutes 31.1 seconds | 9 minutes 10.8 seconds | 24 minutes 22.0 seconds | 1 minute 24.3 seconds | |||
3-21 Basis Set Optimisation
Item Value Threshold Converged?
Maximum Force 0.000092 0.000450 YES
RMS Force 0.000015 0.000300 YES
Maximum Displacement 0.001099 0.001800 YES
RMS Displacement 0.000160 0.001200 YES
Predicted change in Energy=-5.666743D-08
Optimization completed.
-- Stationary point found.
6-31G(d,p)Basis Set Optimisation
Item Value Threshold Converged? Maximum Force 0.000030 0.000450 YES RMS Force 0.000007 0.000300 YES Maximum Displacement 0.001664 0.001800 YES RMS Displacement 0.000353 0.001200 YES Predicted change in Energy=-1.729211D-08 Optimization completed.
6-31G(d,p)Basis Set Frequency Calculation
Low frequencies --- -4.9042 -2.0431 -0.0010 -0.0009 -0.0009 5.0633 Low frequencies --- 91.6470 153.9751 211.4258
Item Value Threshold Converged? Maximum Force 0.000030 0.000450 YES RMS Force 0.000007 0.000300 YES Maximum Displacement 0.001664 0.001800 YES RMS Displacement 0.000353 0.001200 YES Predicted change in Energy=-1.729211D-08 Optimization completed.
[N(CH3)3CH2OH]+
| [N(CH3)3CH2OH]+ | |||||||
| Molecule |
| ||||||
|---|---|---|---|---|---|---|---|
| File Type | .log | .log | .log | .log | |||
| Log File | File:NOH1log 70296.log | File:NOH2log 70298.log | File:NOH3log 70302.log | File:NOH4log 70305.log | |||
| D-Space | DOI:10042/22855 | DOI:10042/22856 | DOI:10042/22857 | DOI:10042/22859 | |||
| Calculation Type | FOPT | FOPT | FREQ | SP | |||
| Calculation Method | RB3YLP | RB3YLP | RB3YLP | RB3YLP | |||
| Basis Set | 3-21 | 6-31G(d,p) | 6-31G(d,p) | 6-31G(d,p) | |||
| Final Energy | -287.8066 a.u. | -289.3947 a.u. | -289.3947 a.u. | -289.3947 a.u. | |||
| Gradient | 0.00000631 a.u. | 0.00002696 a.u. | 0.00000529 a.u. | - a.u. | |||
| Dipole Moment | 2.2674 Debye | 2.1353 Debye | 2.1353 Debye | 2.1353 Debye | |||
| CPU Time | 6 minutes 3.1 seconds | 11 minutes 5.3 seconds | 9 minutes 53.8 seconds | 1 minutes 42.6 seconds | |||
3-21 Basis Set Optimisation
Item Value Threshold Converged?
Maximum Force 0.000090 0.000450 YES
RMS Force 0.000021 0.000300 YES
Maximum Displacement 0.001386 0.001800 YES
RMS Displacement 0.000440 0.001200 YES
Predicted change in Energy=-8.835078D-08
Optimization completed.
-- Stationary point found.
6-31G(d,p)Basis Set Optimisation
Item Value Threshold Converged? Maximum Force 0.000022 0.000450 YES RMS Force 0.000005 0.000300 YES Maximum Displacement 0.001142 0.001800 YES RMS Displacement 0.000261 0.001200 YES Predicted change in Energy=-1.010997D-08 Optimization completed.
Comparison
| Molecule | [N(CH3)4]+ | [N(CH3)3CH2OH]+ | [N(CH3)33CH2CN]+ | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
| |||||||||
| Charge Distribution (+0.8 TO -0.8) | 
|

|

| ||||||||

|

|

| |||||||||
| Bonds | C-H 63% from C (sp3 (26% s, 74%p), 37% from H (100% s) | C-H 63% from C (sp3 (26% s, 74%p), 36% from H (100% s) | C-H 64% from C (sp3 (27% s, 73%p), 36% from H (100% s) | ||||||||
| C-N 34% from C (sp3 (20% s, 80%p)), 66% from N (sp3 (25% s, 75%p)) | C-N 34% from C (sp3 (21% s, 79%p)), 66% from N (sp3 (25% s, 75%p)) | C-N 33% from C (sp3 (20% s, 80%p)), 67% from N (sp3 (25% s, 75%p)) | |||||||||
| HOMO | 
|

|

| ||||||||
| LUMO | 
|

|

| ||||||||
| HOMO E (a.u.) | -0.57933 | -0.48766 | -0.50048 | ||||||||
| HOMO-LUMO GAP E (a.u.) | -0.44631 | -0.36308 | -0.31863 |
| Molecule | Atom | Charge |
|---|---|---|
| [N(CH3)4 ]+ | N | -0.295 |
| C | -0.483 | |
| H | 0.269 | |
| [N(CH3)3CH2OH]+ | N | 0.726 |
| C | -0.511 | |
| (CH2)H | 0.259-0.284 | |
| C(OH)HH | 0.237 | |
| O | -0.678 | |
| OH | 0.493 | |
| [N(CH3)33CH2CN]+ | N | 0.411 |
| C | 0.194-0.208 | |
| (CH2)H | 0.184-0.284 | |
| CCN | -0.089 | |
| C(CN)HH | 0.220 | |
| CCN | 0.354 | |
| N | -0.394 |
Comparison
The Homos and lumos are very different for the substituted compounds. With the unsubstituted compound, [N(CH3)4]+ the HOMO and LUMO are spread fairly evenly throughout the molecule, particularly the LUMO, which displays loose Td symmetry. However, when the molecule has different substituents on one of the methyl groups, the LUMO and HOMO change accordingly. With an OH substituent, the HOMO becomes a lot more compact. This can be expected as the oxygen donates electrons to the rest of the molecule. Coupled with the electronegativity of the N, the homo is mainly focused on these two molecules and the bridging carbon. The LUMO however is very diffuse, with the carbons and hetero atoms in phase and the hydrogens out of phase; which expands out from the structure of the molecule. The LUMO of the -CN substituted molecule follows a similar pattern, with the heteroatoms all being in phase and the hydrogens (plus half of the CN) and all the space in between is out of phase. However it is not as diffuse as the OH substituted molecule as CN is an electron withdrawing group. The HOMO of the -CN molecule is almost exclusively focused on the cyano group and bridging carbon. This can be rationalised as the CN withdraws electrons away from the rest of the molecule. With the addition of a functional group, the MOs tend to focus around the functional group due to the effect they have on the distribution of electrons within the molecule (either EWG for CN or EDG for O). The HOMOs for both molecules are focused ont he functional groups. The addition of a functional group has raised the energy of the HOMO in both cases here, and decreased the size of the HOMO-LUMO gap; this implies that both of these molecules are more reactive than their unfunctionalised analogue.
Conclusion
After initially optimising and analysing a small molecule, five different cations where optimised and analysed to determine how the charge is distributed amongst the constituent atoms. While qualitative MO theory was shown to have a high level of accuracy, this further analyses showed that some traditional pictures of the location of charges are inaccurate.
References
- ↑ M. Schuurman, W. Allen, H. Schaefer, Journal of Computational Chemistry, 2005, 26, 1106
- ↑ Landau LD and Lifshitz EM (1976) Mechanics, 3rd. ed., Pergamon Press. ISBN 0-08-021022-8 (hardcover) and ISBN 0-08-029141-4 (softcover)
- ↑ The Electrochemical Flow Capacitor: A New Concept for Rapid Energy Storage and Recovery, Volker Presser, Christopher R. Dennison, Jonathan Campos1, Kevin W. Knehr, Emin C. Kumbur2, Yury Gogotsi1, <DOI|10.1002/aenm.201100768>