Rep:Mod:bb407
Inorganic Computational Chemistry Lab
Vibrational Analysis
Vibrational analysis calculations are a function of the potential energy surface (PES) with respect to equilibrium geometry's bond length. The particular function used is a second derivative.
Using this calculation, the output provided information regarding the optimised structure. The optimisation can give an indication of whether the ground or transitional states are most stable. The output also allows for the analysis of the structure's vibrational frequencies.
By analysing the vibrational frequencies, information on the structure can be deduced. Positive frequencies throughout the output data indicate an optimised geometry in the ground state. A single negative frequency in amongst the positive, is an indication of a transitional state. Multiple negative frequencies means that the molecular structure needs further optimisation.
In the following exercise, the optimisation must take place prior to vibrational analysis. The model used was the RB3LYP calculation. A basis set has to be used in conjunction with the calculation. The basis set that was chosen was the LANL2MB. This set was used as the calculations involve optimising a molecular structure that contains chlorine.
As a second row atom, chlorine needed the more complex basis set to accomodate its electronic structure.
BCl3
|
Diagram |
3D Representation |
|||
|---|---|---|---|---|
|
Optimised Structure
|
B-Cl Distance (Å) |
1.87 |
|---|---|
|
Bond Angle (Degrees) |
120 |
Optimisation Calculation
|
File Type |
.log |
|---|---|
|
Calculation Type |
FOPT |
|
Calculation Method |
RB3LYP |
|
Basis Set |
LANL2MB |
|
Energy (a.u.) |
-69.44 |
|
Dipole Moment (D) |
0.00 |
|
Point Group |
D3H |
|
Time (s) |
21.0 |
Frequency Calculation
|
File Type |
.log |
|---|---|
|
Calculation Type |
FREQ |
|
Calculation Method |
RB3LYP |
|
Basis Set |
LANL2MB |
|
Energy (a.u.) |
-69.44 |
|
Dipole Moment (D) |
0.00 |
|
Point Group |
D3H |
|
Time (s) |
17.0 |
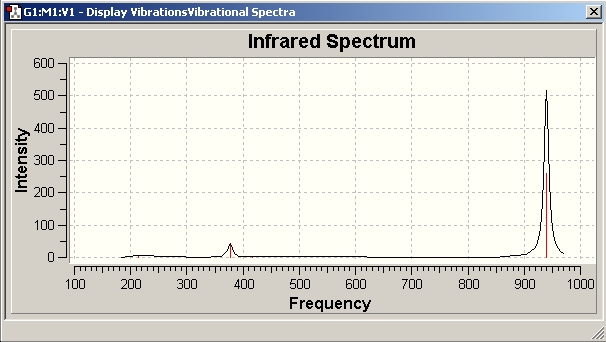
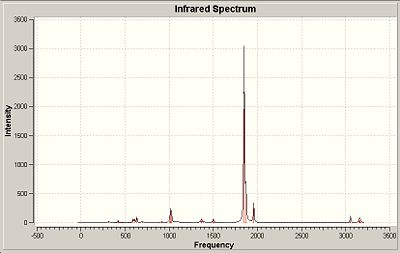
BCl3 Vibrational Analysis
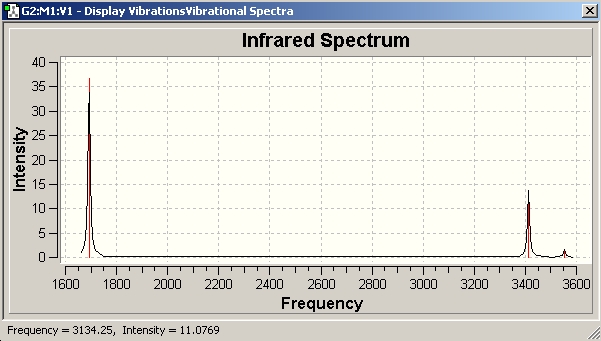
The data provides an IR spectra from which further anaysis was made.
|
Vibration Diagram |
Intensity |
Frequency (cm-1) |
Symmetry: D3H |
Vibration Desciption |
|---|---|---|---|---|
|
258.67 |
939.47 |
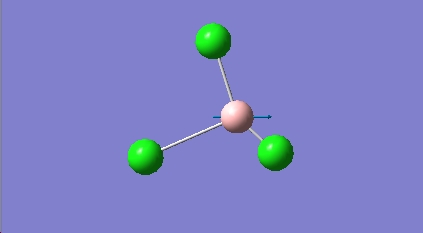
Assymmetric Stretch E' |
Boron oscillates along B-Cl bond |
|
|
258.67 |
939.47 |
Assymmetric Stretch E' |
Boron oscillates perpendicular to plane of molecule |
|
|
0.00 |
417.38 |
Symmetric Stretch A1' |
Chlorine atoms oscillate in time along B-Cl bonds |
|
|
43.78 |
376.94 |
Assymmetric Stretch A2 |
Boron oscillates perpendicular to plane of molecule; Slight movement of Chlorine atoms |
|
|
3.94 |
214.13 |
Assymmetric Bend E' |
Two B-Cl bonds scissor in plane; Remaining B-Cl oscillates along bond |
|
|
3.94 |
214.13 |
Assymmetric Bend E' |
Single B-Cl oscillates in plane of molecule |
On inspection of the data it is apparent that the vibrational frequencies are all positive. This means that the molecule is at a ground state. The mathematical implication is that the molecule has reached a nadir for the bond length with respect to PES.
As expected, the molecule has the correct number of vibrational frequencies. This is based on the vibrational modes afforded to non-linear molecules; 3n-6. Simple spectroscopic theory provides the rationale to interpret the data. Dipole moments provide the activity in infra-red spectroscopy. The symmetric A1' stretch produces no change in dipole moment, and as a result is inactive.
Degenerate E' sets have a difference as a result of variance between strecthing and bending. The B-Cl bending vibration is located at 214.13 cm-1; B-Cl stretch vibrations at 939.47 cm-1. The intensity difference again correlates to stretching having a greater change than the bending vibrations.
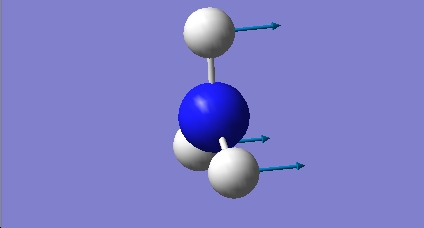
H2O
For the optimisation of this molecule, which is composed of much simpler component atoms than BCl3, the 3-21G basis set was used in conjunction with the RB3LYP method. A simpler basis set is sufficient to adequately model the electronic structure.
|
Diagram |
3D Representation |
|||
|---|---|---|---|---|
|
Optimised Structure
|
B-Cl Distance (Å) |
1.00 |
|---|---|
|
Bond Angle (Degrees) |
104.0 |
Optimisation Calculation
|
File Type |
.log |
|---|---|
|
Calculation Type |
FOPT |
|
Calculation Method |
RB3LYP |
|
Basis Set |
3-21G |
|
Energy (a.u.) |
-75.97 |
|
Dipole Moment (D) |
2.24 |
|
Point Group |
C2V |
|
Time (s) |
21.0 |
Frequency Calculation
|
File Type |
.log |
|---|---|
|
Calculation Type |
FREQ |
|
Calculation Method |
RB3LYP |
|
Basis Set |
3-21G |
|
Energy (a.u.) |
-75.97 |
|
Dipole Moment (D) |
2.24 |
|
Point Group |
C2V |
|
Time (s) |
16.0 |
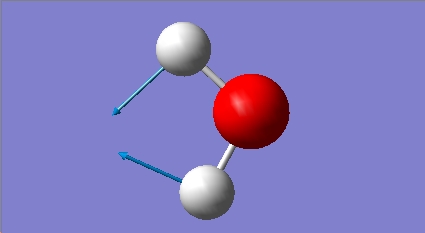
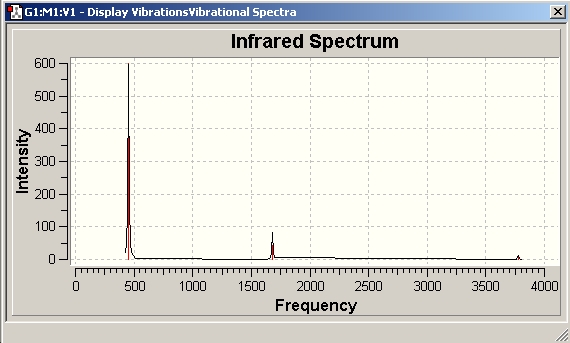
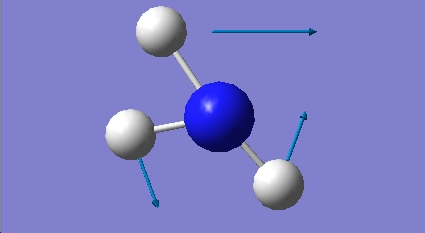
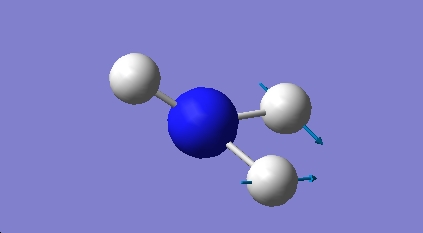
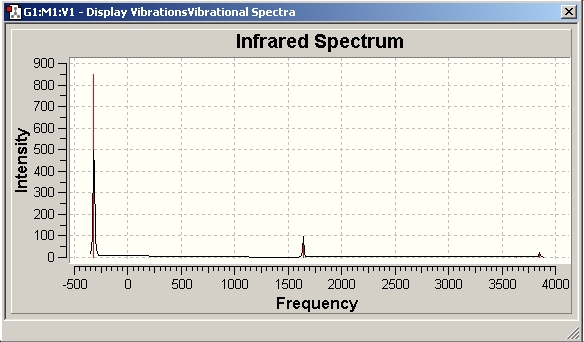
H2O Vibrational Analysis
The data provides an IR spectra from which further anaysis was made.
|
Vibration Diagram |
Intensity |
Frequency (cm-1) |
Symmetry: C2V |
Vibration Desciption |
|---|---|---|---|---|
|
1.56 |
3553.34 |
Assymmetric Stretch B1 |
Oxygens oscillate out-of-time to each other along O-H bonds |
|
|
13.71 |
3412.40 |
Assymmetric Stretch A1 |
Oxygens oscillate in-time with each other along O-H bonds |
|
|
36.74 |
1692.47 |
Bend A1' |
O-H bonds move in a scissoring action |
On inspection of the data it is apparent that the vibrational frequencies are all positive. This means that the molecule is at a ground state. The mathematical implication is that the molecule has reached a nadir for the bond length with respect to PES.
As expected, the molecule has the correct number of vibrational frequencies. This is based on the vibrational modes afforded to non-linear molecules; 3n-6.
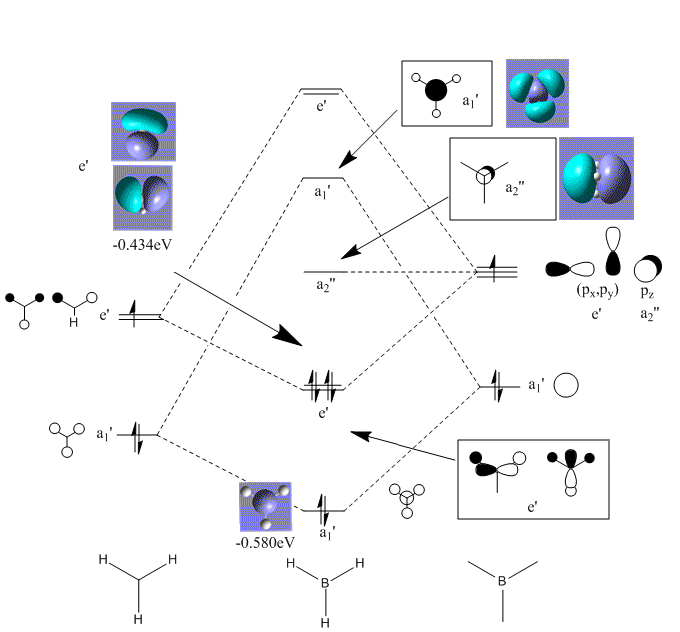
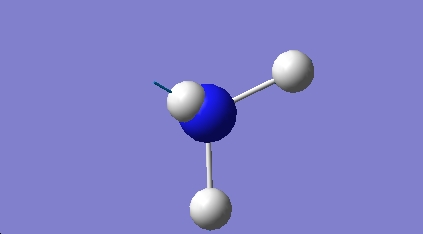
BH3 Molecular Orbital Analysis
Using Gaussview a Molecular Orbital (MO) diagram was put together. The example used was BH3, which was covered during previous lecture courses.
The MO diagram produced is very helpful in providing a qualitative arguement for reactivity. In order for BH3 to complete its octet, it requires two more electrons. It is a well known fact that BH3 is lewis acidic. The a2" orbital shows that the MO occupies a large and broad area above and below the plane of the molecule, allowing for easy attack from a lewis base.
Interestingly, Gaussview predicted four filled MOs. However, the omitted MO was significantly lower in energy and so left out on the basis that this was an artefact of the prediction process.
NH3
In this section, ammonia is to be studied as it possesses the ability to tunnel between PES minima. This means that the structure can invert in a process that superficially is not dissimilar to the inversion on Sn2 centres. This provides two equivalent mirror states with C3V symmetry and a transition state with D3H.
To investigate further the RB3LYP calculation was used again in conjunction with the 6-31G basis set.
NH3 Optimisation Calculation
|
File Type |
.log |
|---|---|
|
Calculation Type |
FOPT |
|
Calculation Method |
RB3LYP |
|
Basis Set |
6-31G |
|
Energy (a.u.) |
-56.53 |
|
Dipole Moment (D) |
1.32 |
|
Point Group |
C3V |
|
Time (min) |
1:14 |
NH3 (Long Bond) Optimisation Calculation
In order to perform this optimisation, symmetry was ignored while one of the N-H bonds was given a set distance of 1.01Å.
This produces a variance in symmetry and in the computing time that was required. The energy difference between the long bond optimisation and the normal optimisation is insignificant.
|
File Type |
.log |
|---|---|
|
Calculation Type |
FOPT |
|
Calculation Method |
RB3LYP |
|
Basis Set |
6-31G |
|
Energy (a.u.) |
-56.53 |
|
Dipole Moment (D) |
1.34 |
|
Point Group |
C1 |
|
Time (min) |
2:20 |
There was a significant increase in calculation time. This has been attributed to the methods by which Gaussview runs the calculation. Without the symmetry elements the calculations take longer as the program cannot use symmetry to 'shortcut' the process.
NH3 (High Symmetry) Optimisation Calculation
By using another atom as a 'dummy' the geometry of the molecular structure is forced to maintain a specific chosen shape.
The structure is now in D3H symmetry, explaining the speedy calculation time.
|
File Type |
.log |
|---|---|
|
Calculation Type |
FOPT |
|
Calculation Method |
RB3LYP |
|
Basis Set |
6-31G |
|
Energy (a.u.) |
-56.53 |
|
Dipole Moment (D) |
0.00 |
|
Point Group |
D3H |
|
Time (min) |
0:31 |
NH3 (RMP2-FU) Optimisation Calculation
In the following optimisation calculations the RMP2-FU method was used in conjunction with the 6-311+G(d,p) basis set. The method and basis set should provide a better calculation.
|
File Type |
.log |
|---|---|
|
Calculation Type |
SP |
|
Calculation Method |
RMP2-FU |
|
Basis Set |
6-311+G(d,p) |
|
Energy (a.u.) |
-56.43 |
|
Dipole Moment (D) |
1.66 |
|
Point Group |
C3V |
|
Time (min) |
0:16 |
NH3 (RMP2-FU High Symmetry) Optimisation Calculation
|
File Type |
.log |
|---|---|
|
Calculation Type |
FOPT |
|
Calculation Method |
RMP2-FU |
|
Basis Set |
6-311+G(d,p) |
|
Energy (a.u.) |
-56.43 |
|
Dipole Moment (D) |
0.00 |
|
Point Group |
D3H |
|
Time (min) |
0:47 |
On inspection of all optimisation results; the RB3LYP method provides the lowest energy with the long bond structure. However, the variance between all the energy figures is insignificant.
The barrier to inversion is calculated by taking the difference in energy between the C3V and D3H forms.
By taking the values from the above optimisations, the barrier to conversion was calculated to be 19.21 kJmol-1. This is comparable to the literature value of 24.3 kJmol-1.
NH3 (C3V Symmetry) Vibrational Analysis
|
Vibration Diagram |
Intensity |
Frequency (cm-1) |
Symmetry: C3V |
Vibration Desciption |
|---|---|---|---|---|
|
7.09 |
3775.76 |
Asymmetric Stretch E |
Two Hydrogen atoms oscillate out-of-time with each other; Remaining Hydrogen stationary |
|
|
7.09 |
3775.76 |
Asymmetric Stretch E |
Two hydrogen atoms oscillate together and the other out-of-time along the bonds |
|
|
0.07 |
3575.43 |
Symmetric Stretch A1' |
Hydrogen atoms oscillate in time along N-H bonds |
|
|
41.72 |
1680.47 |
Bend E |
Two N-H bonds scissor in plane; Remaining B-Cl oscillates in plane side-to-side |
|
|
41.73 |
1680.47 |
Bend E |
Two N-H bonds scissor in plane; Remaining B-Cl still |
|
|
599.47 |
452.30 |
Bend A1 |
Nitrogen oscillates perpendicular to the plane |
The results have all positive frequencies showing a ground state structure.
NH3 (D3H Symmetry) Vibrational Analysis
|
Vibration Diagram |
Intensity |
Frequency (cm-1) |
Symmetry: D3H |
Vibration Desciption |
|---|---|---|---|---|
|
19.60 |
3854.27 |
Asymmetric Stretch E' |
Two Hydrogen atoms oscillate in-time with each other; Remaining Hydrogen out-of-time |
|
|
19.60 |
3854.27 |
Asymmetric Stretch E<'/P> |
Two Hydrogen atoms oscillate out-of-time with each other; Remaining Hydrogen stationary |
|
|
0.00 |
3653.71 |
Symmetric Stretch A1' |
Hydrogen atoms oscillate in time along N-H bonds |
|
|
55.98 |
1640.59 |
Bend E' |
Two N-H bonds scissor in plane; Remaining B-Cl oscillates in plane side-to-side |
|
|
55.98 |
1640.59 |
Bend E' |
Two N-H bonds scissor in plane; Remaining B-Cl still |
|
|
849.113 |
-318.05 |
- |
Nitrogen oscillates perpendicular to the plane |
With a single negative frequency value, the structure is a transition state.
Transition Metal Complex
In this investigation, a transition metal complex will be analysed via its vibrational frequency data. The cis and trans isomers of the Mo(CO)4(PMe3)2complex will be examined.
The structure is much more complex than the previous examples, and as such requires extra effort to optimise. Initially a RB3LYP calculation is completed using the LANL2MB basis set. Then further optimisation using the LANL2-DZ basis set.
Trans Isomer
The Trans Isomer

|
File Type |
.log |
|---|---|
|
Calculation Type |
FOPT |
|
Calculation Method |
RB3LYP |
|
Basis Set |
LANL2DZ |
|
Energy (a.u.) |
-773.36 |
|
Dipole Moment (D) |
0.00 |
|
Point Group |
C1 |
|
Time (h) |
3:32:42.0 |
The optimisation provides the following data:
|
Optimised Calculation |
Literature Value |
|
|---|---|---|
|
Mo-P Bond |
2.029Å |
2.426Å |
|
Mo-C Bond |
2.029Å, 2.029Å |
2.037Å, 2.041Å |
|
P-Mo-P Angle (Degrees) |
180.0 |
176.7 |
|
R-P-Mo (Degrees) |
117.09, 117.06, 115.68 |
121.02, 120.52, 111.56 |
Cis Isomer
The Cis Isomer
|
File Type |
.log |
|---|---|
|
Calculation Type |
FOPT |
|
Calculation Method |
RB3LYP |
|
Basis Set |
LANL2DZ |
|
Energy (a.u.) |
-773.36 |
|
Dipole Moment (D) |
9.07 |
|
Point Group |
C1 |
|
Time (h) |
6:15:21.6 |
The optimisation provides the following data:
|
Optimised Calculation |
Literature Value |
|
|---|---|---|
|
Mo-P Bond |
2.648Å, 2.648Å |
2.525Å, 2.533Å |
|
Mo-C Bond |
1.982Å, 1.982Å, 2.033Å |
1.981Å, 1.983Å, 2.034Å |
|
P-Mo-P Angle (Degrees) |
92.9 |
94.8 |
|
R-P-Mo (Degrees) |
113.90, 118.26, 118.54 |
112.9, 114.9, 121.1 |