Rep:Mod:banana
This Wiki page presents several different molecules with the formula EX3 in its first part and then looks at the properties of the four isomers of the dimer formed by AlCl2Br, comparing them to each other and also to the monomer. In all cases, the molecular structure is examined and the bonds compared; the Vibrational Spectra are also presented and compared to both other Spectra and also the raw data that shows active and inactive modes.
A bond is an interaction between two atoms. Strong interactions can arise through the sharing of electrons (covalent bonds) or an electrostatic force attracting one another (ionic bonds), whereas weaker interactions arise from bond types such as hydrogen bonding and also dipole interactions. In the case of covalent bonding, the overlap of atomic orbitals (to form a molecular orbital) is the main factor in determining the strength of the bond and so atoms with similar size orbitals will form stronger bonds than those with very different sized bonds. An example of a strong bond is the triple bond in N₂, with an energy of approximately 942KJ/mol, whereas a relatively weak bond such as hydrogen bonding in water can be as low as 23.3KJ/mol. Medium strength bonds, such as a single bond between two carbons, lie between these two extremes – 346KJ/mol in this case.
Although any interaction between two atoms can be defined as a bond of some sort, Gaussview does not always draw bonds where they may be expected or where there is an interaction. This is because when originally programmed, pre-defined distances from experimental data/literature were inputted as the limit to which Gaussview should draw in a bond; if the bond is longer than the literature length it will not appear, even though it is expected to exist.
On this page, for each optimisation, a table with a summary of the optimisation, the confirmation from the .log file that the molecule has been optimised and a jmol image is presented as raw data. For each frequency calculation, a table with the calculation summary and the list of low frequecies is presented, followed by a table of vibrations and the calculated spectrum itself.
EX3 Summary and Comparison
Geometry
| BH3 | BBr3 | GaBr3 | |
|---|---|---|---|
| r(E-X) | 1.20Å | 1.93Å | 2.35Å |
| θ(X-E-X) | 120.0° | 120.0° | 120.0° |
All three molecules optimised have the same bond angle of 120°. This is as expected for an EX₃ molecule as this is the optimal bond angle to minimise repulsive interactions between bonds. What is different between the three molecules however are the bond lengths.
The B-H bond length is very close to the sum of the atomic radii of the two molecules, suggesting a very good overlap of orbitals and the formation of a fairly strong bond. Changing the ligand from H to Br sees an extremely large increase in molecular energy. This is because the orbital overlap is much better as the atoms are more similar sizes. The bond length does remain much longer than in BH₃ though because of this repulsive force between ligands.
When the Boron is replaced with Gallium, the total energy of the molecule is actually lower than in BBr₃. This is because the Gallium atom is larger than the Boron, and so the bromines are able to spread themselves out further from each other giving more diffuse Molecular orbitals and reducing the interactions between them. It is worth noting though that the bond lengths in GaBr₃ and BBr₃ are not as different when compared to the Hydrogen ligand, suggesting that the ligand has a larger effect on the bond length than the central atom.
Vibrational Spectra and Frequency Analyses
Frequency analyses are carried out in order to confirm that the energy of the molecule is at a minimum rather than at a transition state. Optimisations only look for turning points in the Potential Energy surface (i.e. where the gradient/first derivative equals zero), so it is important to check that the point is a minima rather than a maxima. To do this, the second derivative is looked at to check that the gradient is increasing and this is what the frequency analysis looks at. Providing that the low frequencies are zero or positive (within a 15cm⁻¹ tolerance in the case of these calculations), the molecule is at an energy minima and so has been correctly optimised.
When comparing the Vibrational Spectra of molecules, it is important that the frequency analyses have been run using the same basis set. This is because each basis set models the molecule to different models/degrees of accuracy, meaning that each will inherently give different results for the same molecule, let alone when trying to compare different molecules. The “Low Frequencies” that are produced by the frequency analyses represent the movement of the central atom in the molecule/the movement around the molecules central point – in a fully optimised molecule these should be very close to zero.
The frequency of the vibration of a bond/bonds is given by the equation:
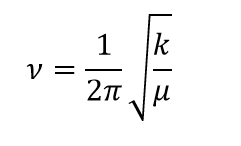
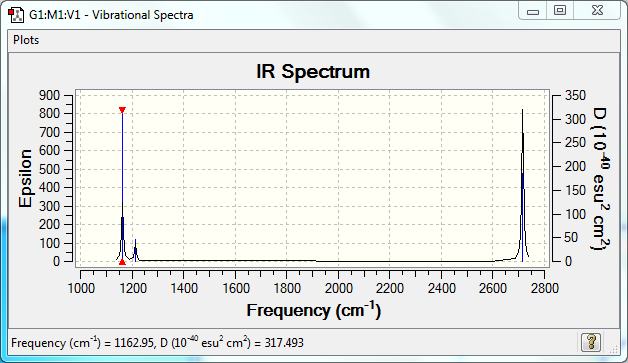
Where "k" is the force constant and "μ" is the reduced mass of the molecule. The frequencies of the BH3 modes range from 1163cm-1 to 2582cm-1; the face that the reduced mass of the molecule will be very small suggests that the force constant and so the bond strength of the B-H bond is very stron - in line with what teh calculated data has indicated.
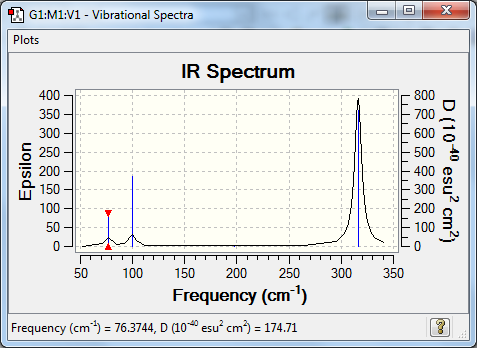
In GaBr3 however, the frequencies range from 76cm-1 to 316cm-1, which is a range of much lower values. This is due to the fact that the reduced mass value has increased significantly and so even if the force constant/bond strength was stronger, it has not compensated enough to maintain the frequency values. The fact that the frequency is only 1/10th to 1/3rd of the value rather than even less suggests that the force contant - and thus the bond strength - has increased slightly; this is backed up by the molecular energy values from the respective optimisations.
It is also worth noting that in BH3, the lowest frequency in the spectrum belongs to the movement of the three ligands simultaneously up and down (with the central atom oscillating in the opposite direction). However in GaBr3, this frequency is in the middle of the spectrum at 100cm-1, suggesting that this oscillation has a stronger force constant relative to others; perhaps a refleection of the good orbital overlap in the Ga-Br bonds.
BH3
Optimisation to B3YLP/3-21G Level
The log file for this optimisation can be found here.
Optimisation to B3YLP/6-31G(d,p) Level
The log file for this optimisation can be found here.
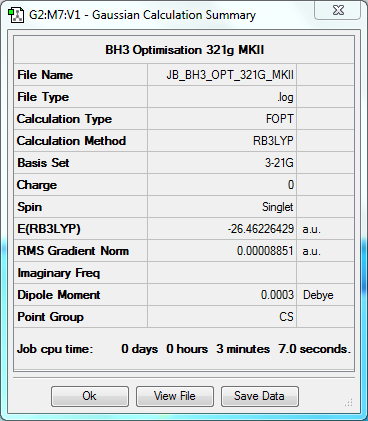
| Data Summary | Convergence | Jmol | |||
|---|---|---|---|---|---|
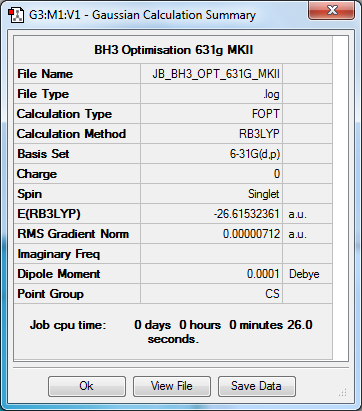
|
Item Value Threshold Converged? Maximum Force 0.000012 0.000450 YES RMS Force 0.000008 0.000300 YES Maximum Displacement 0.000063 0.001800 YES RMS Displacement 0.000039 0.001200 YES |
|
Symmetry Optimisation to D3h at B3YLP/6-31G(d,p) Level
The log file for this optimisation can be found here.
| Data Summary | Convergence | Jmol | |||
|---|---|---|---|---|---|
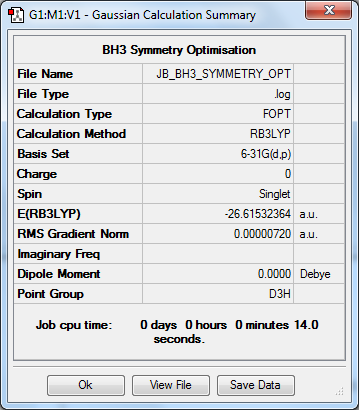
|
Item Value Threshold Converged? Maximum Force 0.000014 0.000450 YES RMS Force 0.000009 0.000300 YES Maximum Displacement 0.000057 0.001800 YES RMS Displacement 0.000037 0.001200 YES |
|
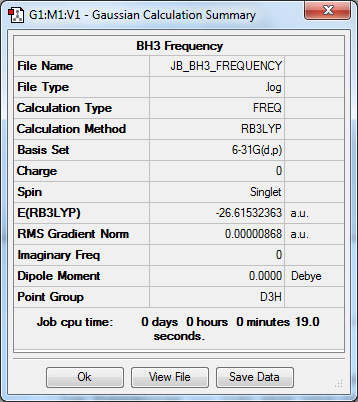
Frequency to B3YLP/6-31G(d,p) Level
The frequency file can be found here.
| Data Summary | Low Modes |
|---|---|
|
Low frequencies --- -10.1111 -3.0903 -0.0054 0.4892 1.8274 3.5901 Low frequencies --- 1162.9534 1213.1537 1213.1564 |
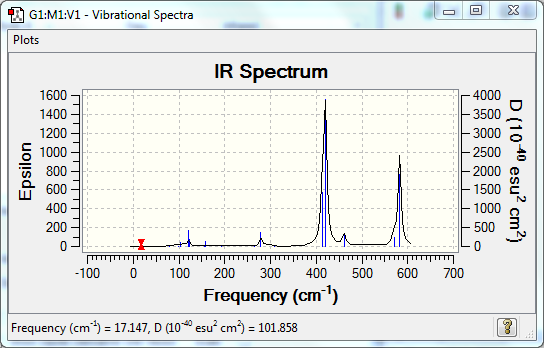
Vibrational Spectrum
| Wavenumber | Intensity | IR active? | Type |
| 1163 | 93 | yes | bend |
| 1213 | 14 | very slight | bend |
| 1213 | 14 | very slight | bend |
| 2582 | 0 | no | stretch |
| 2716 | 126 | yes | stretch |
| 2716 | 126 | yes | stretch |
MO Population at B3YLP/6-31G(d,p) Level
The log file for this calculation can be found here. DOI:10042/31225

As shown by the image above, there are strong similarities between the LCAO-calculated orbitals and those calculated by Gaussian. This adds weight to the argument that this method is a valid way of quickly calculating Molecular Orbitals without the need for a powerful computer.
BBr3
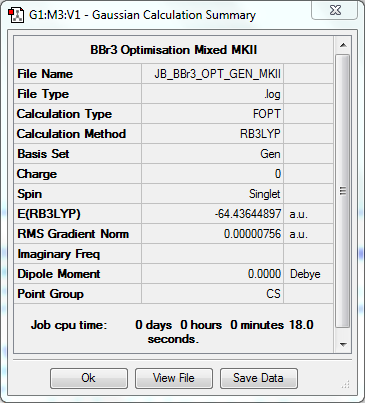
Optimisation to B3YLP/LANL2DZ Level for Br, 6-31G(d,p) for B
The log file for this optimisation can be found here. DOI:10042/31199
| Data Summary | Convergence | Jmol | |||
|---|---|---|---|---|---|
|
Item Value Threshold Converged? Maximum Force 0.000016 0.000450 YES RMS Force 0.000009 0.000300 YES Maximum Displacement 0.000090 0.001800 YES RMS Displacement 0.000059 0.001200 YES |
|
GaBr3
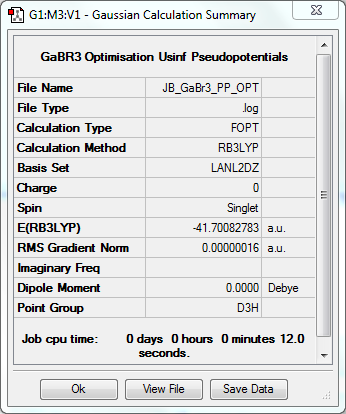
Optimisation to B3YLP/LANL2DZ Level
The log file for this optimisation can be found here. DOI:10042/31198
| Data Summary | Convergence | Jmol | |||
|---|---|---|---|---|---|
|
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000003 0.001800 YES RMS Displacement 0.000002 0.001200 YES |
|
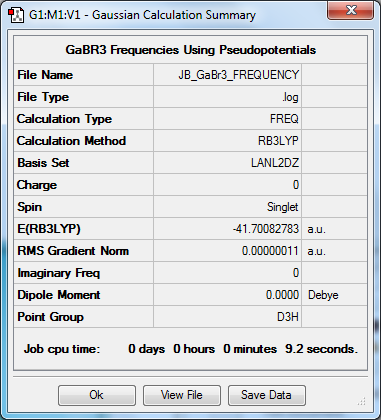
Frequency to B3YLP/LANL2DZ Level
The frequency file can be found here. DOI:10042/31221
| Data Summary | Low Modes |
|---|---|
|
Low frequencies --- -0.5252 -0.5247 -0.0024 -0.0010 0.0235 1.2010 Low frequencies --- 76.3744 76.3753 99.6982 |
Vibrational Spectrum
| Wavenumber | Intensity | IR active? | Type |
| 76 | 3 | very slight | bend |
| 76 | 3 | very slight | bend |
| 100 | 9 | very slight | bend |
| 197 | 0 | no | stretch |
| 316 | 57 | yes | stretch |
| 316 | 57 | yes | stretch |
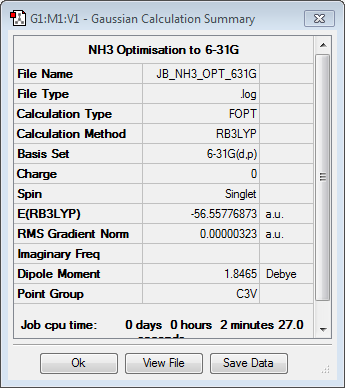
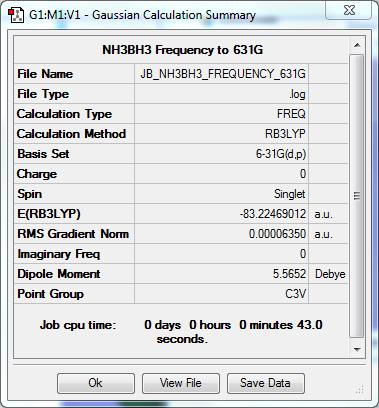
NH3
Optimisation to B3YLP/6-31G(d,p) Level
The log file for this optimisation can be found here.
| Data Summary | Convergence | Jmol | |||
|---|---|---|---|---|---|
|
Item Value Threshold Converged? Maximum Force 0.000006 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000012 0.001800 YES RMS Displacement 0.000008 0.001200 YES |
|
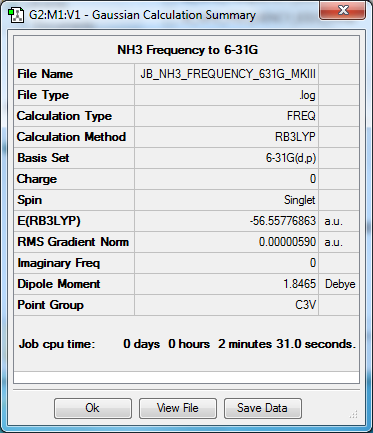
Frequency to B3YLP/6-31G(d,p) Level
The frequency file can be found here.
| Data Summary | Low Modes |
|---|---|
|
Low frequencies --- -8.5223 -8.4750 -0.0025 0.0335 0.1919 26.4067 Low frequencies --- 1089.7616 1694.1862 1694.1866 |
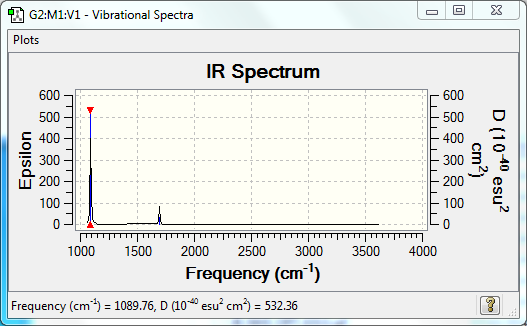
Vibrational Spectrum
| Wavenumber | Intensity | IR active? | Type |
| 1090 | 145 | yes | bend |
| 1694 | 14 | very slight | bend |
| 1694 | 14 | very slight | bend |
| 3461 | 1 | no | stretch |
| 3589 | 0 | no | stretch |
| 3589 | 0 | no | stretch |
MO Population at B3YLP/6-31G(d,p) Level
The log file for this calculation can be found here. DOI:10042/31245
| Calculated HOMO for NH3 | Calculated LUMO for NH3 |
|---|---|

|

|
NBOs at B3YLP/6-31G(d,p) Level
The image below shows the charge distribution within an NH3 molecule, with bright red being highly negative (-1.125) and bright green highly positive (+1.125). In this case, the Nitrogen atom has a charge of -1.125 and the Hydrogen atoms each have a charge of +0.375.

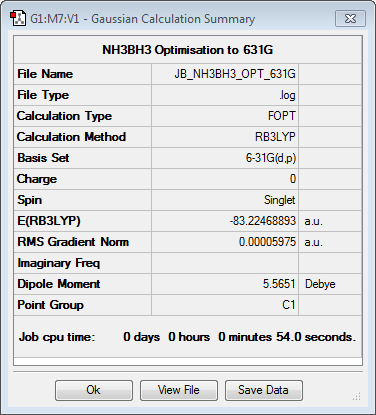
BH3NH3
Optimisation to B3YLP/6-31G(d,p) Level
The log file for this optimisation can be found here.
| Data Summary | Convergence | Jmol | |||
|---|---|---|---|---|---|
|
Item Value Threshold Converged? Maximum Force 0.000122 0.000450 YES RMS Force 0.000058 0.000300 YES Maximum Displacement 0.000513 0.001800 YES RMS Displacement 0.000296 0.001200 YES |
|
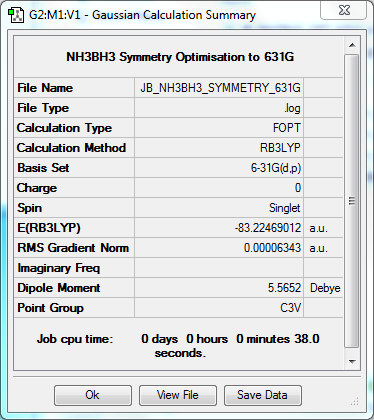
Symmetry Optimisation to C3v at B3YLP/6-31G(d,p) Level
The log file for this optimisation can be found here.
| Data Summary | Convergence | Jmol | |||
|---|---|---|---|---|---|
|
Item Value Threshold Converged? Maximum Force 0.000121 0.000450 YES RMS Force 0.000059 0.000300 YES Maximum Displacement 0.000605 0.001800 YES RMS Displacement 0.000315 0.001200 YES |
|
By taking the molecule energies from BH₃ and NH₃ and comparing them to that of BH₃NH₃, it is possible to compute the estimated energy for the B-N bond. This dative covalent bond is expected to be of medium strength, and indeed when calculated, the number is 140KJ/mol (to the nearest 10kj/mol; the approximate accuracy of the Gaussian calculation). This figure is actually slightly larger than what has been measured elsewhere, but it is roughly what would be expected for a bond of this strength. It is worth noting that because the basis set used isn’t perfect, the figure will inherently be slightly off the true value.
Frequency to B3YLP/6-31G(d,p) Level
The frequency file can be found here.
| Data Summary | Low Modes |
|---|---|
|
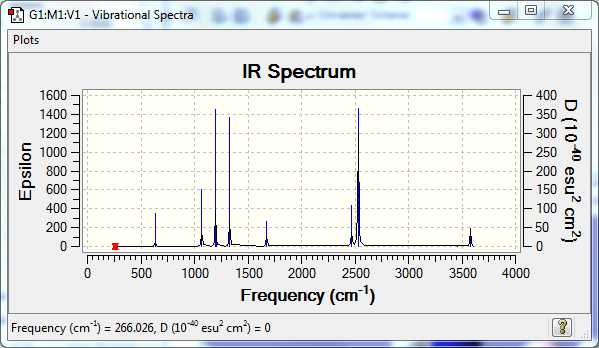
Low frequencies --- -0.0614 -0.0456 -0.0064 21.6984 21.7043 40.6154 Low frequencies --- 266.0421 632.3708 640.1454 |
Vibrational Spectrum
| Wavenumber | Intensity | IR active? | Type |
| 266 | 0 | no | twist |
| 632 | 14 | very slight | stretch |
| 640 | 4 | no | bend |
| 640 | 4 | no | bend |
| 1069 | 41 | yes | bend |
| 1069 | 41 | yes | bend |
| 1197 | 109 | yes | bend |
| 1204 | 3 | no | bend |
| 1204 | 3 | no | bend |
| 1330 | 114 | yes | bend |
| 1677 | 28 | yes | bend |
| 1677 | 28 | yes | bend |
| 2470 | 67 | yes | stretch |
| 2530 | 231 | yes | stretch |
| 2530 | 231 | yes | stretch |
| 3463 | 3 | no | stretch |
| 3579 | 28 | yes | stretch |
| 3579 | 28 | yes | stretch |
Project - Lewis Acids and Bases
All raw data, vibrational frequency tables and spectra can be found after this section. The aim of this mini-project is to analyse and compare the four different dimers formed when AlCl2Br dimerises and also to compare them to the monomer unit.
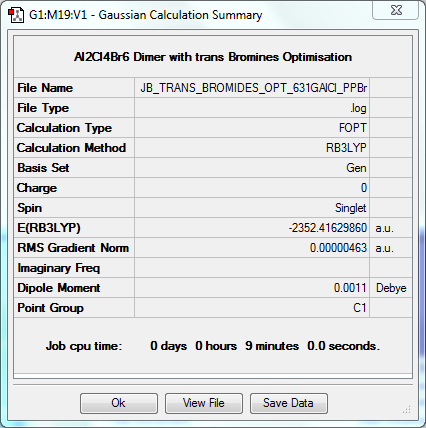
Using data from the calculations carried out, it can be determined that using these basis sets, the most stable dimer of AlCl₂Br is the arrangement where the two bromine atoms are terminal and arranged in what is effectively a trans-position to one another (Isomer 3). Comparing this to 2 monomer units, the dissociation energy can be calculated as 0.03135 a.u., or 80KJ/mol; a surprisingly large value which confirms that the monomer is less stable than the dimer.
The isomer with two bridging Bromines (isomer 1) is the least stable of the four isomers. This is because while the Bromine atoms are a more similar size to Aluminium, when arranged in a four membered ring, they are large enough to cause unfavourable interactions, lowering the stability by 0.00998 a.u. or 26KJ/mol and thus making this isomer very unlikely to spontaneously occur.
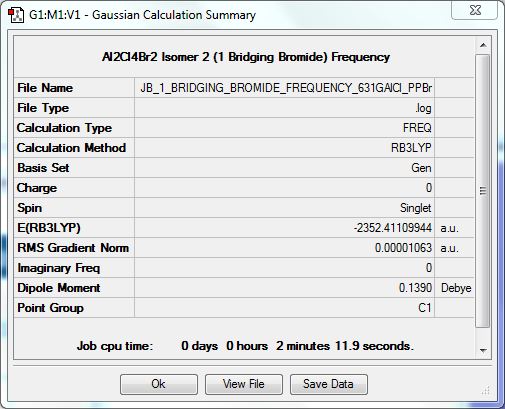
When the number of bridging Bromines is reduced to one (i.e. the other is a terminal Bromine; Isomer 2), the molecule is a bit less destabilised compared to the molecule with two as there are less destabilising interactions between the Bromines, though there is now more strain on the ring as the smaller Cl atom makes it slightly smaller. In this case, the molecule is destabilised by 0.00518 a.u. or 14KJ/mol.
The isomers with two terminal Bromines “cis” (Isomer 4) and “trans” (Isomer 3) relative to each other are extremely similar as at this point the two Bromine atoms both have very good overlap with their Aluminium atoms and are far enough away from each other to avoid almost any unfavourable interactions. This “cis” isomer is 0.00002 a.u. or 0.1KJ/mol less stable than the “trans” conformation. This value is just due to the fact that the Bromines are even further away from each other in the latter; even though the Bromines are still a long distance apart in the “cis” isomer, there are still some tiny interactions. Thus it is probably reasonable to expect a mix of “cis” and “trans” isomers when dimerisation occurs.
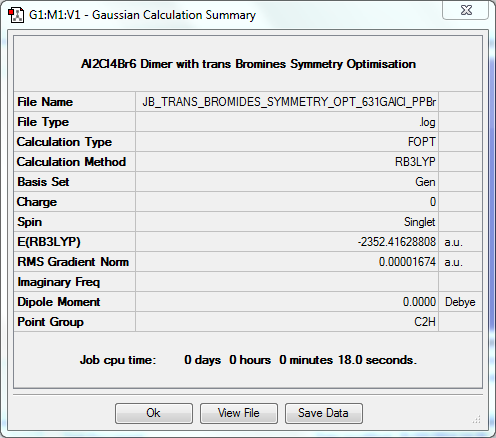
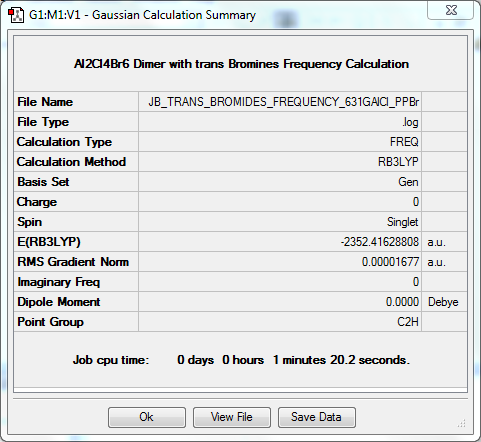
In order for a bend, stretch or twist to be IR active, there must be a net dipole moment i.e. the action must not cancel itself out. When looking with these four isomers, it is clear that there is a strong correlation between the degree of symmetry in the isomer and the number of active IR vibrations. There are much fewer active vibrations in Isomer 1 and Isomer 3, where the degree of symmetry is greater. Isomer 2 has many more active vibrations as its point group is C₁, and Isomer 4 has slightly more than Isomer 1 and 3, but less than Isomer 4 as it is still slightly more symmetric.
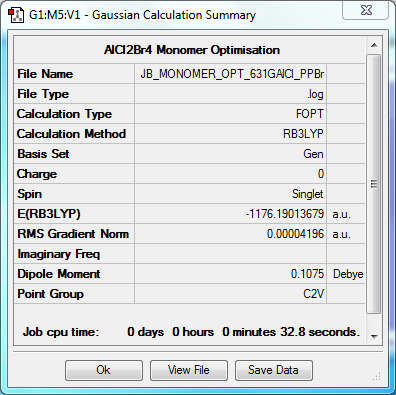
Monomer
Optimisation to B3YLP/6-31G(d,p)for Al and Cl, LanLD2Z for Br
The log file for this optimisation can be found here. DOI:10042/31293
| Data Summary | Convergence | Jmol | |||
|---|---|---|---|---|---|
|
Item Value Threshold Converged? Maximum Force 0.000136 0.000450 YES RMS Force 0.000073 0.000300 YES Maximum Displacement 0.000681 0.001800 YES RMS Displacement 0.000497 0.001200 YES |
|
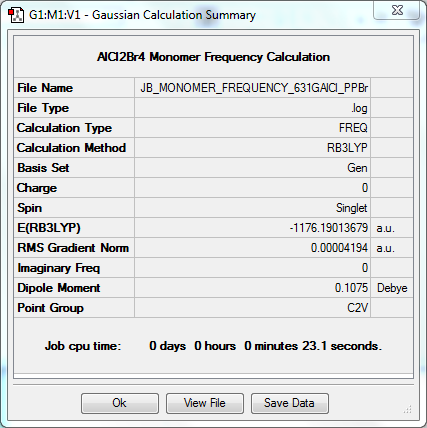
Frequency Calculation
The frequency file can be found here. DOI:10042/31362
| Data Summary | Low Modes |
|---|---|
|
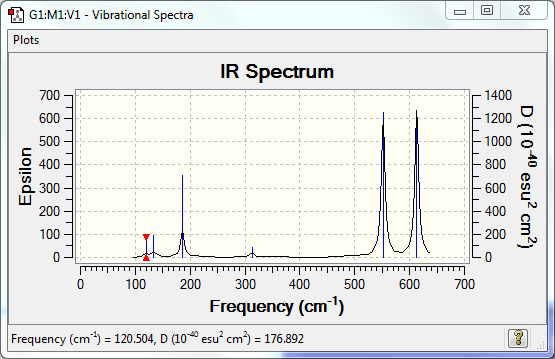
Low frequencies --- -0.0014 -0.0009 0.0025 1.3569 3.6367 4.2604 Low frequencies --- 120.5042 133.9178 185.8950 |
Vibrational Spectrum
| Wavenumber | Intensity | IR active? | Type |
| 121 | 5 | very slight | bend |
| 134 | 6 | very slight | bend |
| 186 | 33 | yes | bend |
| 313 | 7 | very slight | stretch |
| 552 | 174 | yes | stretch |
| 613 | 186 | yes | stretch |
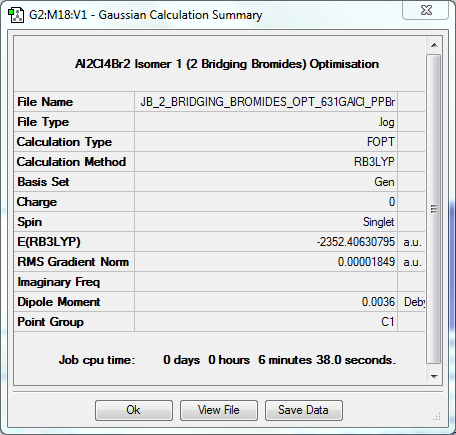
Dimer Isomer 1: 2 Bridging Bromides
Optimisation to B3YLP/6-31G(d,p)for Al and Cl, LanLD2Z for Br
The log file for this optimisation can be found here. DOI:10042/31252
| Data Summary | Convergence | Jmol | |||
|---|---|---|---|---|---|
|
Item Value Threshold Converged? Maximum Force 0.000046 0.000450 YES RMS Force 0.000011 0.000300 YES Maximum Displacement 0.000927 0.001800 YES RMS Displacement 0.000371 0.001200 YES |
|
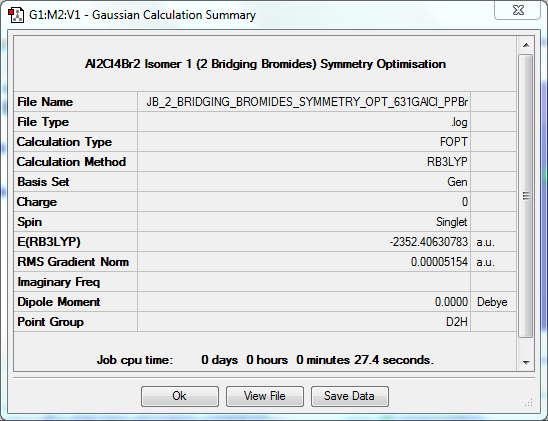
Symmetry Optimisation to D2h
The log file for this optimisation can be found here. DOI:10042/31294
| Data Summary | Convergence | Jmol | |||
|---|---|---|---|---|---|
|
Item Value Threshold Converged? Maximum Force 0.000101 0.000450 YES RMS Force 0.000037 0.000300 YES Maximum Displacement 0.000585 0.001800 YES RMS Displacement 0.000231 0.001200 YES |
|
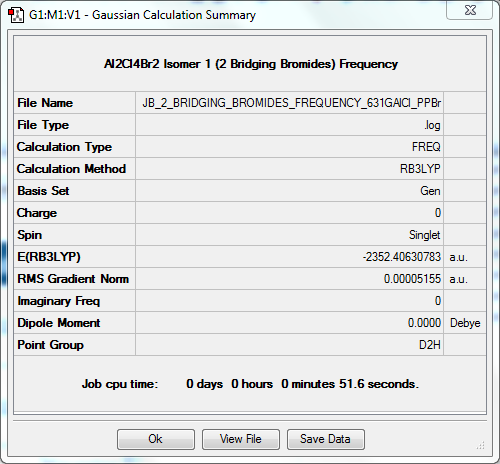
Frequency Calculation
The frequency file can be found here. DOI:10042/31298
| Data Summary | Low Modes |
|---|---|
|
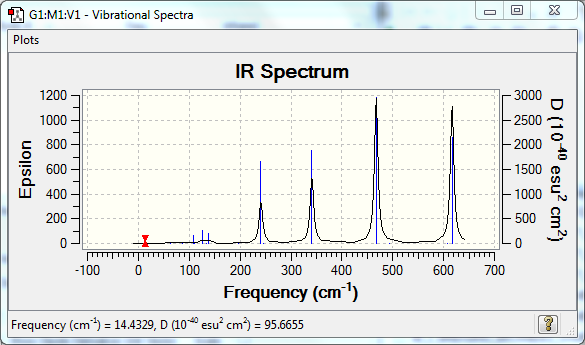
Low frequencies --- -6.0061 -5.7551 -3.3098 0.0029 0.0035 0.0037 Low frequencies --- 14.4329 63.0991 85.9935 |
Vibrational Spectrum
| Wavenumber | Intensity | IR active? | Type |
| 14 | 0 | no | bend |
| 63 | 0 | no | twist |
| 86 | 0 | no | bend |
| 87 | 1 | no | bend |
| 107 | 5 | no | bend |
| 111 | 0 | no | bend |
| 126 | 8 | very slight | bend |
| 134 | 0 | no | twist |
| 138 | 7 | very slight | bend |
| 162 | 0 | no | bend |
| 197 | 0 | no | stretch |
| 241 | 100 | yes | stretch |
| 247 | 0 | no | stretch |
| 341 | 160 | yes | stretch |
| 468 | 347 | yes | stretch |
| 494 | 0 | no | stretch |
| 608 | 0 | no | stretch |
| 617 | 332 | yes | stretch |
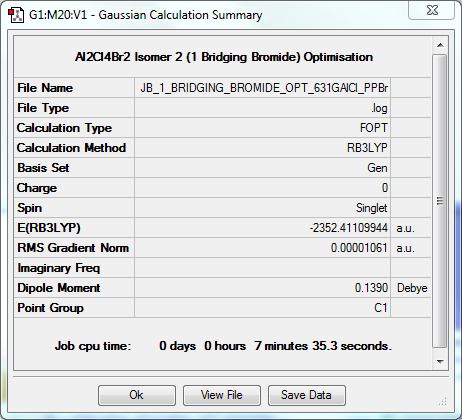
Dimer Isomer 2: 1 Bridging Bromide
Optimisation to B3YLP/6-31G(d,p)for Al and Cl, LanLD2Z for Br
The log file for this optimisation can be found here. DOI:10042/31253
| Data Summary | Convergence | Jmol | |||
|---|---|---|---|---|---|
|
Item Value Threshold Converged? Maximum Force 0.000020 0.000450 YES RMS Force 0.000009 0.000300 YES Maximum Displacement 0.000974 0.001800 YES RMS Displacement 0.000384 0.001200 YES |
|
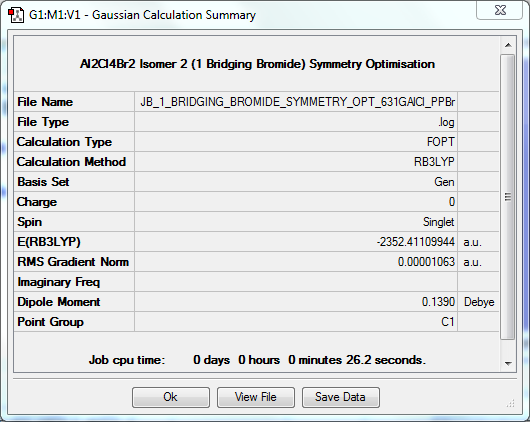
Symmetry Optimisation to C1
The log file for this optimisation can be found here. DOI:10042/31301
| Data Summary | Convergence | Jmol | |||
|---|---|---|---|---|---|
|
Item Value Threshold Converged? Maximum Force 0.000020 0.000450 YES RMS Force 0.000009 0.000300 YES Maximum Displacement 0.000737 0.001800 YES RMS Displacement 0.000272 0.001200 YES |
|
Frequency Calculation
The frequency file can be found here. DOI:10042/31302
| Data Summary | Low Modes |
|---|---|
|
Low frequencies --- -2.4966 -0.0041 -0.0032 -0.0030 0.2773 3.0910 Low frequencies --- 17.1118 55.9235 80.0656 |
Vibrational Spectrum
| Wavenumber | Intensity | IR active? | Type |
| 17 | 0 | no | bend |
| 56 | 0 | no | twist |
| 80 | 0 | no | bend |
| 92 | 1 | no | bend |
| 107 | 0 | no | bend |
| 110 | 5 | no | bend |
| 121 | 8 | very slight | bend |
| 149 | 5 | no | twist |
| 154 | 6 | very slight | bend |
| 186 | 1 | no | bend |
| 211 | 21 | yes | stretch |
| 257 | 10 | slight | stretch |
| 289 | 48 | yes | stretch |
| 384 | 154 | yes | stretch |
| 424 | 274 | yes | stretch |
| 492 | 107 | yes | stretch |
| 574 | 122 | yes | stretch |
| 614 | 197 | yes | stretch |
Dimer Isomer 3: Bromines "Trans" to One Another
Optimisation to B3YLP/6-31G(d,p)for Al and Cl, LanLD2Z for Br
The log file for this optimisation can be found here. DOI:10042/31271
| Data Summary | Convergence | Jmol | |||
|---|---|---|---|---|---|
|
Item Value Threshold Converged? Maximum Force 0.000010 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.001019 0.001800 YES RMS Displacement 0.000408 0.001200 YES |
|
Symmetry Optimisation to C2h
The log file for this optimisation can be found here. DOI:10042/31357
| Data Summary | Convergence | Jmol | |||
|---|---|---|---|---|---|
|
Item Value Threshold Converged? Maximum Force 0.000023 0.000450 YES RMS Force 0.000013 0.000300 YES Maximum Displacement 0.000277 0.001800 YES RMS Displacement 0.000135 0.001200 YES |
|
Frequency Calculation
The frequency file can be found here. DOI:10042/31363
| Data Summary | Low Modes |
|---|---|
|
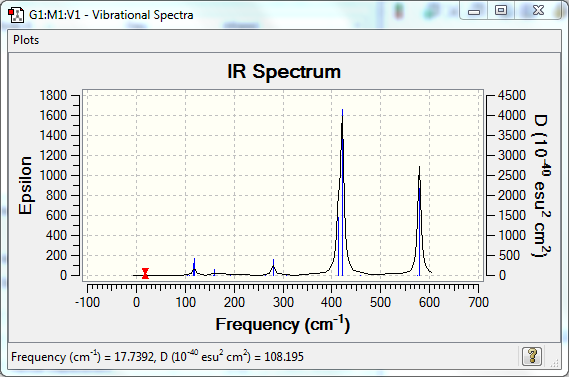
Low frequencies --- -3.9151 -2.1617 -0.0029 -0.0022 0.0019 1.0825 Low frequencies --- 17.7392 48.9866 72.9538 |
Vibrational Spectrum
| Wavenumber | Intensity | IR active? | Type |
| 18 | 0 | no | bend |
| 50 | 0 | no | twist |
| 73 | 0 | no | bend |
| 105 | 0 | no | bend |
| 109 | 0 | no | twist |
| 117 | 9 | very slight | bend |
| 120 | 13 | slight | bend |
| 157 | 0 | very slight | twist |
| 160 | 6 | very slight | bend |
| 192 | 0 | no | bend |
| 264 | 0 | no | stretch |
| 280 | 29 | yes | stretch |
| 308 | 0 | no | stretch |
| 413 | 149 | yes | stretch |
| 421 | 439 | yes | stretch |
| 459 | 0 | no | stretch |
| 574 | 0 | no | stretch |
| 579 | 316 | yes | stretch |
MO Population
The .fchk file used for this calculation can be found here. Molecular Orbitals were generated directly from this file using Gaussview.
Five non-core orbitals were chosen and are presented here alongside an analysis of their bonding/antibonding nature.
| Calculated MO for Isomer 3 | Description |
|---|---|
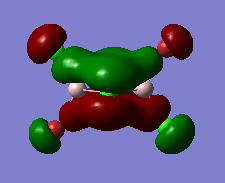
|
Strong bonding interactions across the 4-membered ring, with a planar node in the plane of the ring. All terminal atoms are themselves nodes. Slight antibonding interaction between the orbital density beond the terminal atoms and the ring. All orbitals are not very diffuse and are of similar sizes. |
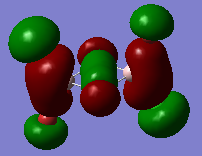
|
Stong bonding interactions between the two bridgins Chlorines. Strong bonding interections also between the terminal atoms and the Aluminium atoms. Nodes are present on all terminal atoms and also the two Aluminium atoms. Net non-bonding between Aluminiums and bridging Chlorines, if anything slightly anti-bonding. Very weak anti-bonding interactions between terminal atoms. All orbitals are of similar size and not diffuse, with those of the terminal Bromine atoms being smaller than those of the terminal Choline atoms. |
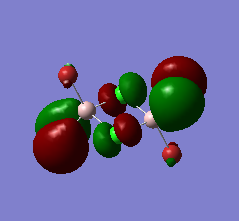
|
Fairy stong anti-bonding interactions between the bridging and terminal Chlorines. Very slight bonding interaction between the terminal Bromines and Cholorines at each end, and very slight anti-bonding between them accross the longer side of the dimer. Nodes can be found on the four terminal atoms, between the terminal atoms on the long sides (above and below the ring) and the two terminal chlorines and bridging chlorines. Very small orbitals on terminal bromines but very diffuse orbitals on terminal cholorines. Bridging chlorines have medium sized orbitals. |
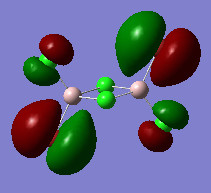
|
Moderate antibonding interactions between all four terminal atoms. No MO density on the 4-membered ring, with nodes on and between all four terminal atoms. The terminal Bromine atoms have very diffuse orbitals, the terminal Chlorine atoms are much smaller. |
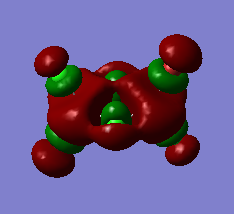
|
Very strong bonding interactions between the two Aluminium atoms and very strong antibonding interactions between them and both the bridging and terminal atoms. Nodes can be found on all bridging and terminal atoms. Aluminium orbitals are diffuse whereas the other orbitals are of a more standard size. The terminal Bromines' orbitals are slightly larger than those of the terminal chlorines. |
Dimer Isomer 4: Bromines "Cis" to One Another
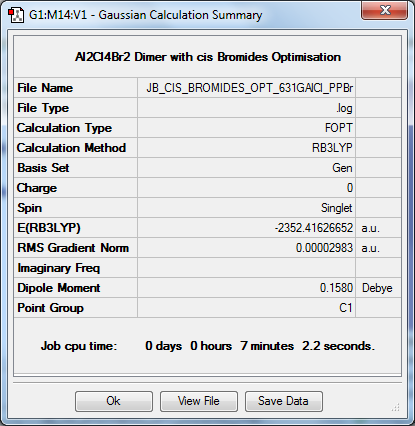
Optimisation to B3YLP/6-31G(d,p)for Al and Cl, LanLD2Z for Br
The log file for this optimisation can be found here. DOI:10042/31280
| Data Summary | Convergence | Jmol | |||
|---|---|---|---|---|---|
|
Item Value Threshold Converged? Maximum Force 0.000058 0.000450 YES RMS Force 0.000025 0.000300 YES Maximum Displacement 0.001668 0.001800 YES RMS Displacement 0.000489 0.001200 YES |
|
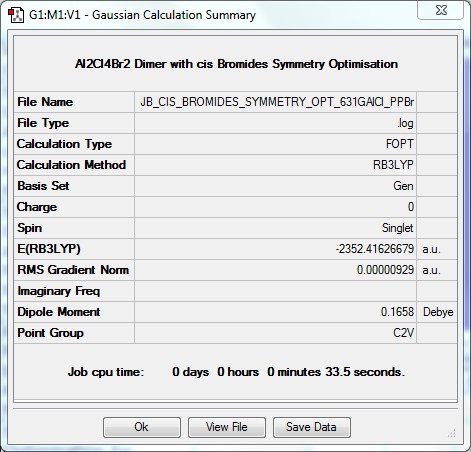
Symmetry Optimisation to C2v
The log file for this optimisation can be found here. DOI:10042/31358
| Data Summary | Convergence | Jmol | |||
|---|---|---|---|---|---|
|
Item Value Threshold Converged? Maximum Force 0.000020 0.000450 YES RMS Force 0.000008 0.000300 YES Maximum Displacement 0.000920 0.001800 YES RMS Displacement 0.000310 0.001200 YES |
|
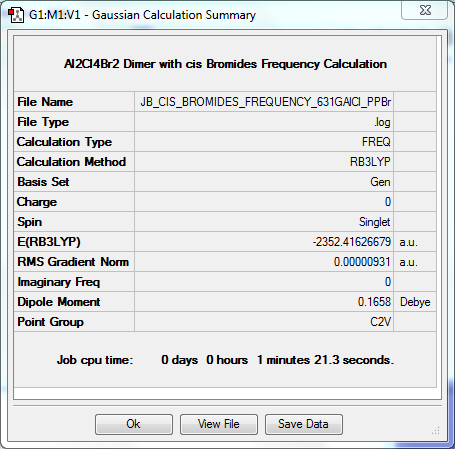
Frequency Calculation
The frequency file can be found here. DOI:10042/31364
| Data Summary | Low Modes |
|---|---|
|
Low frequencies --- -4.4556 -2.4643 -0.0011 -0.0009 -0.0009 1.1640 Low frequencies --- 17.1470 50.8910 78.5536 |
Vibrational Spectrum
| Wavenumber | Intensity | IR active? | Type |
| 17 | 0 | no | bend |
| 51 | 0 | no | twist |
| 79 | 0 | no | bend |
| 99 | 0 | no | bend |
| 103 | 3 | no | bend |
| 121 | 13 | slight | bend |
| 123 | 6 | very slight | bend |
| 157 | 0 | no | twist |
| 158 | 6 | very slight | bend |
| 194 | 2 | no | bend |
| 264 | 0 | no | stretch |
| 279 | 25 | yes | stretch |
| 309 | 2 | no | stretch |
| 413 | 149 | yes | stretch |
| 420 | 411 | yes | stretch |
| 461 | 35 | yes | stretch |
| 570 | 32 | yes | stretch |
| 582 | 278 | yes | stretch |
Project Conclusion
In this project, four different dimers have been modelled alongside the monomer for of AlCl2 and compared. Perhaps unsurprisingly; the molecules with terminal Bromine atoms have proved to be significantly more stable than those with bridging bromides and it has been established that two monomer units are less stable than the dimer form. Further investigation from this point could examine the effects of replacing the Bromine atoms with another Halogen - such as Iodine - and whether the dimer form is still more energetically favorable than the monomer form. It has also been established that the number of active IR frequencies is directly related to the symmetry of the molecule; this is because of the need for a change in dipole moment for the frequency to be IR active, which occurs more frequently in a more asymmetric molecule.
References
Approximate bond energies quoted in the first section of this wiki page were obtained from [1]. The Hydrogen-bonding energy value was obtained from [2]. Some information for analysing the vibrational spectra was obtained [3]. The lab script for this experiment can be found at [4]. The Gaussian Manual also proved to be very useful [5]!
All websites were last accessed 24/10/2014.
All other data was obtained and interpreted via Gaussview. No images from external sources have been used.