Rep:Mod:awc106 module2 DAY2
Day 2: Cis Trans Isomerism
Introduction
In this section we are going to carry out calculations on Mo(CO)4(Me3)2. We will use the calculations performed to predict the stability and spectral data of the two isomers, cis and trans. We will be able to use the CO vibrational bands to determine the symmetry of the two complexes. We expect four bands for the cis isomer, and one band for the trans isomer.
In order to achieve a good result, the optimisations were set up as follows. The cis and trans forms were drawn in ChemBio3D and saved as .gjf files and subsequently opened in Gaussview. A calculation was set up, using the B3LYP method, and a basis set of LANL2MB, with "opt=loose" written in the keywords section. This calculation was used to obtaine a rough initial geometry. The two calculations were sent to the SCAN service to run overnight. Once these calculations had finished, further calculations were run from the optimised geometries, this time using a higher basis set of LANL2DZ and "int=ultrafine scf=conver=9" in the keywords section which increased the electronic convergence. This is a higher basis set and pseudo-potential. These two calculations were again put on the SCAN service overnight.
Trans Mo(CO)4(Me3)2
Table 1: Trans Mo(CO)4(Me3)2 Optimisations | ||||||
| Optimisation | Results | Image | File | |||
| Initial | 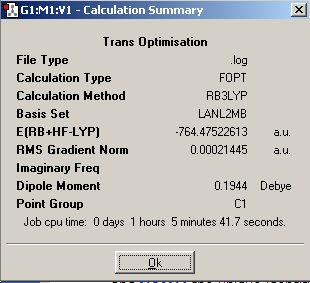
|
|
https://www.ch.ic.ac.uk/wiki/images/4/44/Trans_opt_initial.jpg | |||
| Ultrafine | 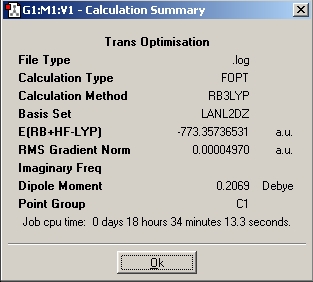
|
|
DOI:10042/to-1798 | |||
| Frequency | 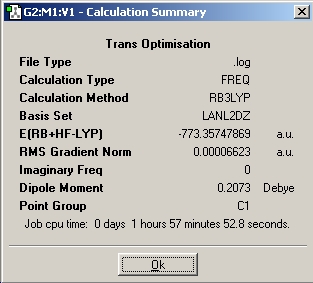
|
|
DOI:10042/to-1801 | |||
Cis Mo(CO)4(Me3)2
Table 1: Cis Mo(CO)4(Me3)2 Optimisations | ||||||
| Optimisation | Results | Image | File | |||
| Initial | 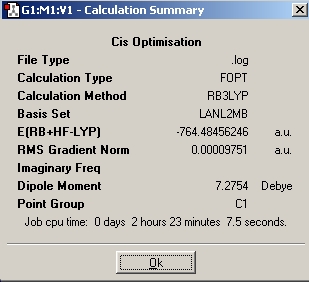
|
|
DOI:10042/to-1723 | |||
| Ultrafine | 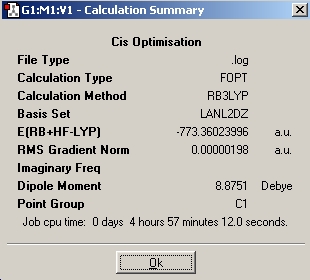
|
|
DOI:10042/to-1799 | |||
| Frequency | 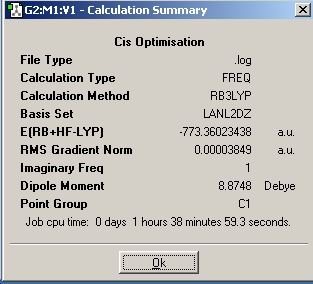
|
|
DOI:10042/to-1800 | |||
compare relevant geometric parameters from the optimised geometries and experimental literature values (it is up to you to find these values).
You don't have to find exactly the same moleucle, as we have used a model to reduce the computational cost! Also depending on your ligand it may be hard to find even a similar molecule. In this case try to use literature values to estimate a rough range for the bond length to be (for example Mo-N single vs Mo=N double bonds). We are looking to see if your computation is a good one, and you haven't made some mistake which has allowed the calculation to complete, but gives rubbish results. This is part of computational chemistry ... always checking that your results make sense, as the computer cannot do that for you. While this is useful it shouldn't take up a large portion of your time, 1-2 hrs max! If you can't find the values you need, tell me how you searched for them and offer suggestions for another way of checking the accuracy of your calculations
compare the computed cis and trans geometries
IR
Trans Spectra

Cis Spectra

Table 1: Mo(CO)4(Me3)2 IR Data | |||
| Trans Frequency/cm-1 | Trans Intensity | Cis Frequency/cm-1 | Cis Intensity |
| 1839.0 | 1999.0 | 1848.5 | 1068.8 |
| 1839.3 | 1999.8 | 1850.7 | 1960.9 |
| 1882.9 | 6.1 | 1869.9 | 666.3 |
| 1954.4 | 3.1 | 1960.4 | 321.7 |
From the information obtinaed, we can calculate the energy difference between the two isomers. We can see that the trans isomer is the more stable with the lower total energy. Can we imagine a scenario in which it was the cis isomer which was more stable? By fine-tuning the molecule, by altering the ligands, we can alter the relative ordering of the two isomers. We can see that the cis isomer is more sterically hindered, thus by reducing the steric bulk of the ligands we can imagine that the energy difference would decrease, and even the possibility of the cis isomer becoming the more stable.
