Rep:Mod:alexou
Introduction to molecular dynamics simulation
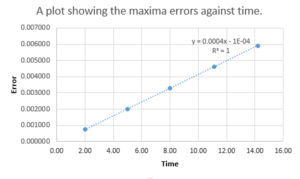
TASK: For the default timestep value, 0.1, estimate the positions of the maxima in the ERROR column as a function of time. Make a plot showing these values as a function of time, and fit an appropriate function to the data. Look at how the energy varies with time.


For a timestep of 0.1, there is a positive correlation between the maxima errors and time as shown from figure 1.
TASK: Experiment with different values of the timestep. What sort of a timestep do you need to use to ensure that the total energy does not change by more than 1% over the course of your "simulation"? Why do you think it is important to monitor the total energy of a physical system when modelling its behaviour numerically?
To ensure that the system is modeled reasonably well, the energy should not change by more than 1%. In this particular case ,this value was found to be 0.2. In real systems the total energy is conserved. In order to simulate and model such systems ,it is important to ensure that the total energy is approximately constant in the simulation to model the system accurately.
TASK: For a single Lennard-Jones interaction, , find the separation, , at which the potential energy is zero. What is the force at this separation? Find the equilibrium separation, , and work out the well depth (). Evaluate the integrals , , and when .
To obtain at which the potential energy is zero from the Lennard-Jones equation,the lennard-Jones potential is set to 0
This is then rearranged for r
and so
,
Hence
Since The force constant at this separation is given by equating the negative differential of the Lennard-Jones potential and setting r to .
when is substituted, the equation is reduced to the following:
This can be further reduced to :
The equilibrium separation, , can be worked out by equating the negative differential of the Lennard-Jones potential to 0.
Rearrange for r
Therefore
The well depth is calculated by substituting for in the Lennard-Jones potential
, , and when .
The following integrals were calculated as -0.0248, -3.29x10^-3, -8/18x10^-4 respectively
TASK: Estimate the number of water molecules in 1ml of water under standard conditions. Estimate the volume of water molecules under standard conditions.
Under standard conditions, 1ml is equal to 1g as the density of water is 1g/1ml. Using the moles equation ,, the moles is calculated as
This is the multiplied by Avogadro constant, 6.023x10^23 to give the number of molecules which is
The volume of water molecules :
TASK: Consider an atom at position in a cubic simulation box which runs from to . In a single timestep, it moves along the vector . At what point does it end up, after the periodic boundary conditions have been applied?.
so
The well depth ,, is equal to
In order to obtain in it is necessary to multiply by Avagadro's constant and divide by 1000.
The reduced temperature can be calculated form the folowing:
Equilibrium
TASK: Why do you think giving atoms random starting coordinates causes problems in simulations? Hint: what happens if two atoms happen to be generated close together?
Problems may arise from providing random atom coordinates even though this would provide a more realistic situation.If two molecule are too positioned too close to each other,the potential of the atoms would be very large and would be converted into kinetic energy, hence this would misrepresent the total energy of the system if many of the molecules are initially positioned very close to each other. Instead, the atoms are placed on the lattice points.
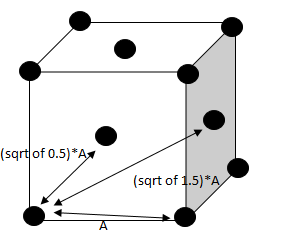
TASK: Satisfy yourself that this lattice spacing corresponds to a number density of lattice points of . Consider instead a face-centred cubic lattice with a lattice point number density of 1.2. What is the side length of the cubic unit cell?
The lattice spacing is determined from the equation below: Numbervdensity = which is equal to: which is approximately 0.8
A rearrangement of the equation above would give:
For a face-centred cubic lattice with a lattice point number density of 1.2, there are 4 particles in the lattice hence :
The side of the cube is equal to
TASK: Consider again the face-centred cubic lattice from the previous task. How many atoms would be created by the create_atoms command if you had defined that lattice instead?
There are 4 atoms in a face centered lattice and 1000 lattices therefore :
1000*4 =4000 atoms are in the system.
TASK: Using the LAMMPS manual, find the purpose of the following commands in the input script:
mass 1 1.0 pair_style lj/cut 3.0 pair_coeff * * 1.0 1.0
mass 1 1.0
The first value,1, represents the atom type The second value,1.0 represents the mass value
pair_style lj/cut 3.0
lj represents the interaction used while /cut 3.0 indicates that any neigbouring atoms greater than 3 will have the potential set to 0.
pair_coeff * * 1.0 1.0
The 1st asterisk represent the atom types, in this case there is only one atom type.
1.0 and 1.0 represent the well depth() and the at which there is no potential between atoms )
TASK: Given that we are specifying and , which integration algorithm are we going to use?
Velocity Verlet Algorithm will be used as and .
TASK: Look at the lines below.
### SPECIFY TIMESTEP ###
variable timestep equal 0.001
variable n_steps equal floor(100/${timestep})
variable n_steps equal floor(100/0.001)
timestep ${timestep}
timestep 0.001
### RUN SIMULATION ###
run ${n_steps}
run 100000
The second line (starting "variable timestep...") tells LAMMPS that if it encounters the text ${timestep} on a subsequent line, it should replace it by the value given. In this case, the value ${timestep} is always replaced by 0.001. In light of this, what do you think the purpose of these lines is? Why not just write:
timestep 0.001 run 100000
This enables it so that the time simulated will be the same for each timestep used but the number of timesteps will be different.
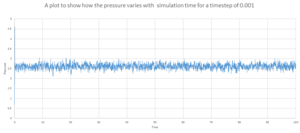
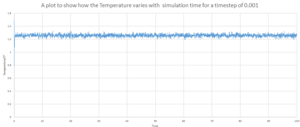
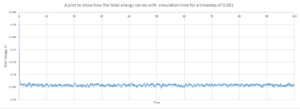
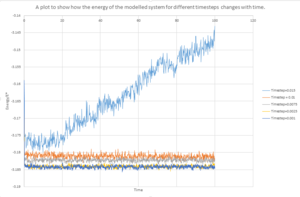
As shown by figure 4-6, they appear to reach equilibrium very quickly after 0.3.
In order to determine the most suitable timestep for simulation used in later parts , It is necessary to consider how well the timestep models the real system and also the amount of simulation time. Smaller timesteps will indefinitely lead to more accurate simulations. However this would only allow the system to be simulated over a short period of time. It can be seen from figure 7 that 4 of the 5 time steps equilibrate very quickly but 0.0075 and 0.01 can be disregarded as this over estimates the energy of the system and appears to have larger fluctuations indicating that the time steps are slightly too large to be used. 0.001 and 0.0025 both essentially overlap indicating that the size of the time step would provide accurate results required in both case and so 0.0025 is chosen as this would provided more simulation time. Also this would require less time to calculate the simulation.
The 0.015 timestep provides the least accurate results and would not be able to simulate the system as the total energy appears to increase with simulation time which is not consistent with the law of conservation as the time step is too large.
Running simulations under specific conditions
TASK: We need to choose so that the temperature is correct if we multiply every velocity . We can write two equations:
Both equations can be added together to give the following equation:
Since E=T
Since
this can be substituted to give :
A further rearrangement and substitution gives:
TASK: Use the manual page to find out the importance of the three numbers 100 1000 100000. How often will values of the temperature, etc., be sampled for the average? How many measurements contribute to the average? Looking to the following line, how much time will you simulate?
Every 100 time steps the input value is used to calculate different functions.The average is calculated every 1000000 steps and the previous 1000 values recorder are used in the average calculation.
TASK: When your simulations have finished, download the log files as before. At the end of the log file, LAMMPS will output the values and errors for the pressure, temperature, and density . Use software of your choice to plot the density as a function of temperature for both of the pressures that you simulated. Your graph(s) should include error bars in both the x and y directions. You should also include a line corresponding to the density predicted by the ideal gas law at that pressure. Is your simulated density lower or higher? Justify this. Does the discrepancy increase or decrease with pressure?
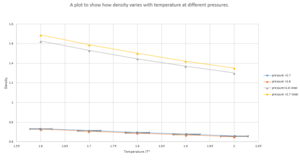
As shown from figure 8, It can be seen that as temperature increases, the density decrease. The simulated densities appear to be lower than the density derived from the ideal gas law. This is because the ideal gas law doesn't account for the interactions between molecules so the simulated data appears to be less dense because the repulsive forces are ignored. At higher pressures the system appears to be more dense which is also consistent as the system is more compact due to higher pressure. The discrepancy between the ideal gas and simulated system for a pressure of 2.7 compared to the 2.6 is higher because of the repulsive forces that occur when molecules are closer together.
The density for the Ideal gas counter part was calculated by the following equation:
where is equal to 1 when reduced.
Calculating heat capacities using statistical physics
TASK: As in the last section, you need to run simulations at ten phase points. In this section, we will be in density-temperature phase space, rather than pressure-temperature phase space. The two densities required at and , and the temperature range is . Plot as a function of temperature, where is the volume of the simulation cell, for both of your densities (on the same graph). Is the trend the one you would expect? Attach an example of one of your input scripts to your report.
### DEFINE SIMULATION BOX GEOMETRY ### lattice sc 0.2 region box block 0 15 0 15 0 15 create_box 1 box create_atoms 1 box
### DEFINE PHYSICAL PROPERTIES OF ATOMS ### mass 1 1.0 pair_style lj/cut/opt 3.0 pair_coeff 1 1 1.0 1.0 neighbor 2.0 bin
### SPECIFY THE REQUIRED THERMODYNAMIC STATE ### variable T equal 2.6 variable timestep equal 0.0025
### ASSIGN ATOMIC VELOCITIES ###
velocity all create ${T} 12345 dist gaussian rot yes mom yes
### SPECIFY ENSEMBLE ###
timestep ${timestep}
fix nve all nve
### THERMODYNAMIC OUTPUT CONTROL ### thermo_style custom time etotal temp press thermo 10
### RECORD TRAJECTORY ### dump traj all custom 1000 output-1 id x y z
### SPECIFY TIMESTEP ###
### RUN SIMULATION TO MELT CRYSTAL ### run 10000 unfix nve reset_timestep 0
### BRING SYSTEM TO REQUIRED STATE ###
variable tdamp equal ${timestep}*100
variable pdamp equal ${timestep}*1000
fix nvt all nvt temp ${T} ${T} ${tdamp}
run 10000
reset_timestep 0
### MEASURE SYSTEM STATE ### unfix nvt fix nve all nve thermo_style custom step etotal temp press density atoms vol variable dens equal density variable dens2 equal density*density variable temp equal temp variable temp2 equal temp*temp variable press equal press variable press2 equal press*press variable totalenergysquare equal etotal*etotal variable totalenergy equal etotal variable volume equal vol fix aves all ave/time 100 1000 100000 v_dens v_temp v_press v_dens2 v_temp2 v_press2 v_totalenergy v_totalenergysquare
run 100000 variable heatcapdidvol equal ((atoms*atoms*(f_aves[8]-(f_aves[7]*f_aves[7]))/(f_aves[2]*f_aves[2]))/vol) variable avedens equal f_aves[1] variable avetemp equal f_aves[2] variable avepress equal f_aves[3] variable errdens equal sqrt(f_aves[4]-f_aves[1]*f_aves[1]) variable errtemp equal sqrt(f_aves[5]-f_aves[2]*f_aves[2]) variable errpress equal sqrt(f_aves[6]-f_aves[3]*f_aves[3])
print "Averages"
print "--------"
print "Density: ${avedens}"
print "Stderr: ${errdens}"
print "Temperature: ${avetemp}"
print "Stderr: ${errtemp}"
print "Pressure: ${avepress}"
print "Stderr: ${errpress}"
print "heatcapacitydvol: ${heatcapdidvol}"
Above is an examples script used to calculate the heat capacity.
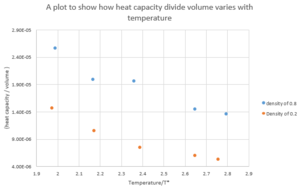
The heat capacity is divided by volume to normalise the heat capacity.As shown from 9,
At higher densities, the molecules are closer on average and hence there is more potential energy in the system which can be converted into kinetic energy. This would mean that the Heat capacity would be higher which is consistent.
Structural properties and the radial distribution function
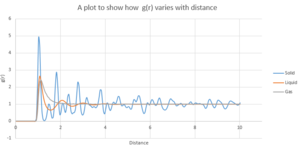
g(r)represents the probability density of finding a particle. For the solid, there are many peaks indicating that there are many areas of high probability indicating many neighbouring particles.This is because the solid has a ordered structure so at a fixed distance there are certain number of particles. Less peaks are seen for the liquid state as the system is less ordered and only one is seen for the gas as the atoms are far apart and the system is more disordered. The troughs indicate the regions where there are lower probability density ("cavities") relative to the density
In all cases there is an distance ,between 0 and 0.5, where there is very little probability. This is a result of repulsion between molecules at small distances. As the distance approaches 10 ,the probability tends to 1 which indicates that there is no predictable arrangement and the probability density is governed by the density of the system.
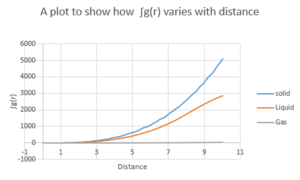
The first 3 peak represent the 3 closest neigbouring particles as shown in figure 12. The closest is followed by then . The lattice spacing can be determined from the 2nd solid peak by reading the distance off the graph which is 1.48 .In order to determine the coordination number,the distance corresponding to the first 3 peaks are taken from the graph and from the figure x,the associated integral (which corresponds to the coordination number). The coordination number for the first 3 peaks are 12 ,18 and 33 respectively.
Dynamical properties and the diffusion coefficient


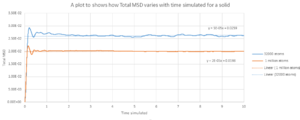
TASK: make a plot for each of your simulations (solid, liquid, and gas), showing the mean squared displacement (the "total" MSD) as a function of timestep. Are these as you would expect? Estimate in each case. Be careful with the units! Repeat this procedure for the MSD data that you were given from the one million atom simulations.
To calculate D, the gradient is multiplied by 1/6.
| Type of simulation | Gradient | D |
|---|---|---|
| 1 million atoms (liquid) | 0.5236 | 0.0872 |
| 1 million atoms (Solid) | 5E-05 | 8.33E-06 |
| 1 million atoms (Gas) | 29.361 | 4.89 |
| 8000 atoms (liquid) | 0.421 | 0.0702 |
| 8000 atoms (Gas) | 15.249 | 2.542 |
| 32000 atoms (Solid) | 2E-05 | 3.33E-06 |
The Diffusion coefficient calculated are appear to be reasonable. As the state changes from solid ,liquid to gas, the coefficient increases. Solid are ordered so we would expect it the value to be close to 0 which is very similar to the value obtained from the simulation where on the other end , the gas would have a larger value then the other 2 states which is true.
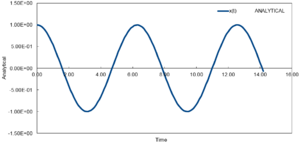
TASK: In the theoretical section at the beginning, the equation for the evolution of the position of a 1D harmonic oscillator as a function of time was given. Using this, evaluate the normalised velocity autocorrelation function for a 1D harmonic oscillator (it is analytic!):
Be sure to show your working in your writeup. On the same graph, with x range 0 to 500, plot with and the VACFs from your liquid and solid simulations. What do the minima in the VACFs for the liquid and solid system represent? Discuss the origin of the differences between the liquid and solid VACFs. The harmonic oscillator VACF is very different to the Lennard Jones solid and liquid. Why is this? Attach a copy of your plot to your writeup.

The minima and maxima represent the points at which the velocity changes direction. For the solid, this is mainly due to the vibrations whereas for the liquid this is due to the collisions with other molecules. In the case for the solid, the decay to 0 is slower compared to the liquid. This is a result of the difference in the order. For the solid ,the structure is fixed and has a greater similarity and so the correlation is higher to-can essentially only vibrate. For the liquid, this is more disordered and will not vibrate but follow a random path so this decays much faster. However, the initial minima is caused by the solvation shell . The Harmonic oscillator is completely different, which resembles a cosine wave, is a result of having no way of transferring the energy. The system can only vibrate -going towards and away form each other which carries on for infinite timesteps as shown in figure 17
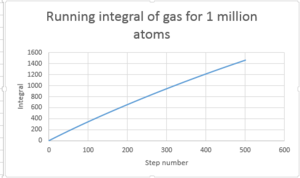
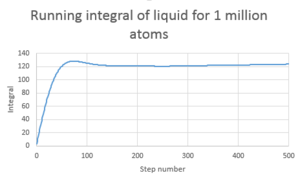
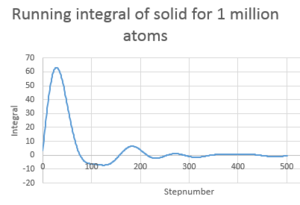
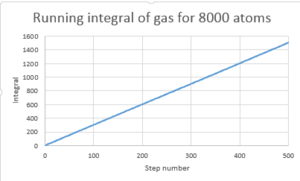
TASK: Use the trapezium rule to approximate the integral under the velocity autocorrelation function for the solid, liquid, and gas, and use these values to estimate in each case. You should make a plot of the running integral in each case. Are they as you expect? Repeat this procedure for the VACF data that you were given from the one million atom simulations. What do you think is the largest source of error in your estimates of D from the VACF?
D in this case is determined by using the trapezium rule and multiplying the value by 1/3
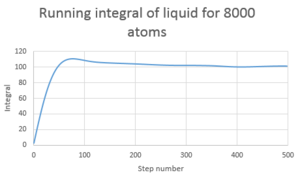
| Type of plot | D |
|---|---|
| Gas 8000atoms | 1000 |
| Gas 1 million atoms | 3299 |
| Solid 32000 atoms | -2.30 |
| Solid 1 million atoms | -0.855 |
| Liquid 8000 atoms | 67.1 |
| Liquid 1 million atoms | 81.9 |
Gas is greatest followed by liquid then solid.

Errors may arise form the fact that only 500 steps are considered and not infinitely as the the line is approximately 0 after 500. The number of atoms can also cause discrepancy. The larger the number atoms the better the system is modeled.