Rep:Mod:aj3318
NH3 molecule
Gaussian calculation summary
Calculation Type FREQ Calculation Method RB3LYP Basis Set 6-31G(d,p) Charge 0 Spin Singlet E(RB3LYP) -56.55776873 a.u. RMS Gradient Norm 0.00000485 a.u. Imaginary Freq 0 Dipole Moment 1.8466 Debye Point Group C3V
Optimisation
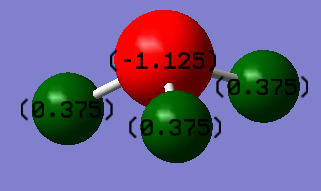
Optimised bond length N-H 1.01798 Å (≈1.02 Å) Optimised bond angle H-N-H 105.741° (≈106°) Charge on N -1.125 D Charge on H 0.375
Item table
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986279D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7412 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7412 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7412 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8571 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Log file
http://wiki.ch.ic.ac.uk/wiki/index.php?title=File:Aj3318_nh3_optf_pop.LOG
Interactive image
NH3 molecule |
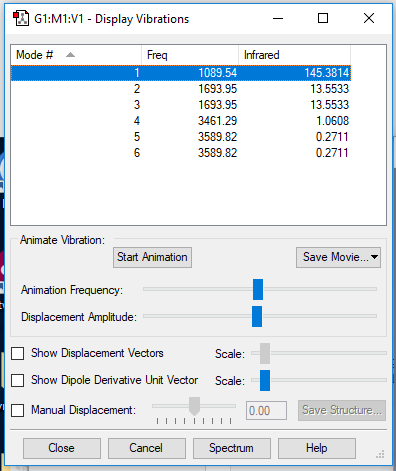
Display Vibrations window
Table of vibrational modes
| wavenumber/cm-1 | 1090 | 1694 | 1694 | 3461 | 3590 | 3590 |
| symmetry | A1 | E | E | A1 | E | E |
| intensity/arb. u. | 145 | 14 | 14 | 1 | 0 | 0 |
| image |  |
 |
 |
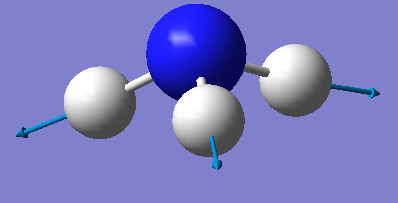 |
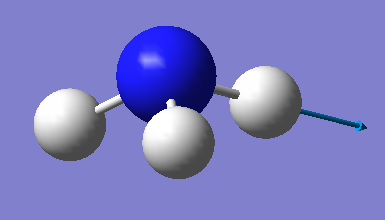 |
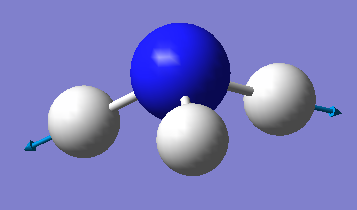
|
Questions
How many modes do you expect from the 3N-6 rule?
6 modes
Which modes are degenerate (ie have the same energy)?
There are two pairs of degenerate E symmetry vibrations. Vibrational mode 2-3, and mode 5-6 are degenerate.
Which modes are "bending" vibrations and which are "bond stretch" vibrations?
Modes 1-3 are bending vibrations and modes 4-6 are stretching vibrations.
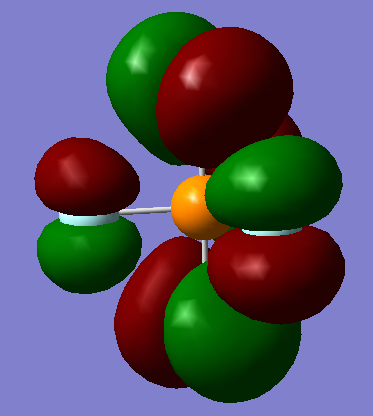
Which mode is highly symmetric?
Mode 1 is highly symmetric - it has C3 symmetry and 3 σv symmetries
One mode is known as the "umbrella" mode, which one is this?
Mode 1
How many bands would you expect to see in an experimental spectrum of gaseous ammonia?
2
Charge distribution NBO
N2 molecule
Gaussian calculation summary
Calculation Type FREQ Calculation Method RB3LYP Basis Set 6-31G(d,p) Charge 0 Spin Singlet E(RB3LYP) -109.52412868 a.u. RMS Gradient Norm 0.00000060 a.u. Imaginary Freq 0 Dipole Moment 0.0000 Debye Point Group D*H
Optimisation
Optimised bond length N-N 1.10550 Å (≈1.11 Å) Charge on N 0 D
Item table
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.400983D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1055 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Log file
https://wiki.ch.ic.ac.uk/wiki/index.php?title=File:AJ3318_N2_OPTF_POP.LOG
Interactive image
N2 molecule |
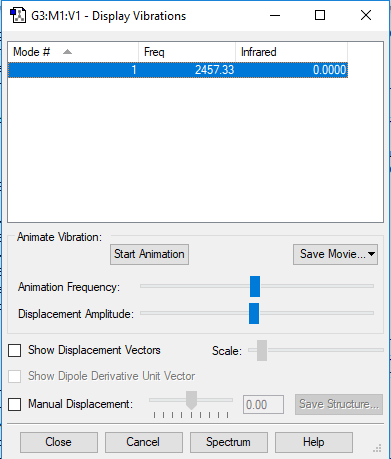
Display Vibrations window
Table of vibrational modes
| wavenumber/cm-1 | 2457 |
| symmetry | SGG |
| intensity/arb. u. | 0 |
| image | 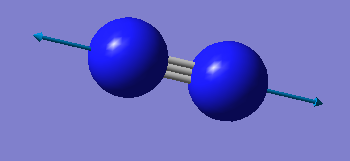
|
Charge distribution NBO
Mono-metallic TM complex
Compound: (dinitrogen)-{2,2',2-(phosphanetriyl)tris[1-(diphenylphosphanyl)-3-methyl-1H-indole]}-ruthenium tetrahydrofuran solvate
Unique identifier: DEKFUX
Link: https://www.ccdc.cam.ac.uk/structures/Search?Ccdcid=DEKFUX&DatabaseToSearch=Published
Reported N-N bond distance: 1.086 Å
Computed N-N bond distance: 1.106 Å
The experimental N#N bond distance in the crystal structure is reported to be smaller than the computed H-H bond distance. This could be because of packing of the structure.
H2 molecule
Gaussian calculation summary
Calculation Type FREQ Calculation Method RB3LYP Basis Set 6-31G(d,p) Charge 0 Spin Singlet E(RB3LYP) -1.17853936 a.u. RMS Gradient Norm 0.00000017 a.u. Imaginary Freq 0 Dipole Moment 0.0000 Debye Point Group D*H
Optimisation
Optimised bond length H-H 0.74279 Å (≈0.74 Å) Charge on H 0 D
Item table
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 0.7428 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Log file
https://wiki.ch.ic.ac.uk/wiki/index.php?title=File:AJ3318_H2_OPTF_POP.LOG
Interactive image
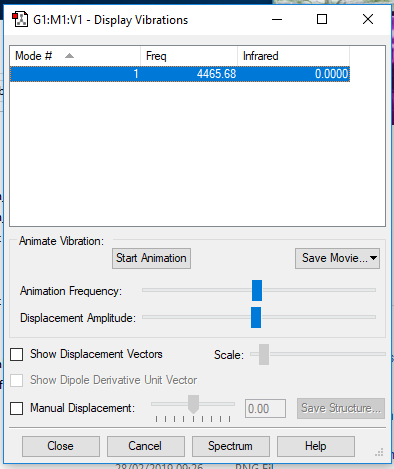
H2 molecule |
Display Vibrations window
Table of vibrational modes
| wavenumber/cm-1 | 4466 |
| symmetry | SGG |
| intensity/arb. u. | 0 |
| image | 
|
Charge distribution NBO
Mono-metallic TM complex
Compound: (s2-Dihydrogen)-tricarbonyl-bis(tri-isopropylphosphine)-tungsten
Unique identifier: CEJDEA
Link: https://www.ccdc.cam.ac.uk/structures/Search?Ccdcid=CEJDEA&DatabaseToSearch=Published
Reported H-H bond distance: 0.755 Å
Computed H-H bond distance: 0.743 Å
The experimental H-H bond distance in the crystal structure is reported to be greater than the computed H-H bond distance. This is because the W-H bond withdraws the electron density, weakening the H-H bond.
Haber-Bosch calculation
E(NH3)= -56.55776873
2*E(NH3)= -113.1155375
E(N2)= -109.52412868
E(H2)= -1.17853936
3*E(H2)= -3.53561808
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]=-0.0557907 au
ΔE=-146.5 kJ mol-1
ΔE<0, so ammonia is more energetically stable than the reacting gas mixtures (H2 and N2).
According to a literature, the standard enthalpy change of formation of ammonia is about -45.6 kJ mol-1. In comparison to the value from computer calculation (-73.2kJ mol-1), this is a significantly lower value. This is probably because computer assumes that all molecules are isolated in infinite space with no vibration/rotation, whereas in practice, intermolecular interactions and molecular kinetics result in overall reduced stabilisation of the ammonia molecules.
Reference: Vanderzee CE, Delbert LK. The enthalpies of solution and formation of ammonia. Journal of Chemical Thermodynamics. 1972, 4(5), 675-683. DOI: 10.1016/0021-9614(72)90039-0
PF5 molecule
Gaussian calculation summary
Calculation Type FREQ Calculation Method RB3LYP Basis Set 6-31G(d,p) Charge 0 Spin Singlet E(RB3LYP) -840.67634601 a.u. RMS Gradient Norm 0.00010182 a.u. Imaginary Freq 0 Dipole Moment 0.0000 Debye Point Group D3H
Optimisation
Optimised bond length P-F 1.56938 Å (≈1.57 Å) Charge on P 2.748 D Charge on F(equatorial) -0.536 D Charge on F(axial) -0.570 D
Item table
Item Value Threshold Converged?
Maximum Force 0.000299 0.000450 YES
RMS Force 0.000090 0.000300 YES
Maximum Displacement 0.000867 0.001800 YES
RMS Displacement 0.000268 0.001200 YES
Predicted change in Energy=-2.765724D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.5694 -DE/DX = -0.0001 !
! R2 R(1,3) 1.5694 -DE/DX = -0.0001 !
! R3 R(1,4) 1.5966 -DE/DX = 0.0003 !
! R4 R(1,5) 1.5694 -DE/DX = -0.0001 !
! R5 R(1,6) 1.5966 -DE/DX = 0.0003 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 90.0 -DE/DX = 0.0 !
! A3 A(2,1,5) 120.0 -DE/DX = 0.0 !
! A4 A(2,1,6) 90.0 -DE/DX = 0.0 !
! A5 A(3,1,4) 90.0 -DE/DX = 0.0 !
! A6 A(3,1,5) 120.0 -DE/DX = 0.0 !
! A7 A(3,1,6) 90.0 -DE/DX = 0.0 !
! A8 A(4,1,5) 90.0 -DE/DX = 0.0 !
! A9 A(5,1,6) 90.0 -DE/DX = 0.0 !
! A10 L(4,1,6,2,-1) 180.0 -DE/DX = 0.0 !
! A11 L(4,1,6,2,-2) 180.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 120.0 -DE/DX = 0.0 !
! D2 D(2,1,5,3) 180.0 -DE/DX = 0.0 !
! D3 D(2,1,6,3) -120.0 -DE/DX = 0.0 !
! D4 D(2,1,5,4) 90.0 -DE/DX = 0.0 !
! D5 D(2,1,6,5) 120.0 -DE/DX = 0.0 !
! D6 D(3,1,5,4) -90.0 -DE/DX = 0.0 !
! D7 D(3,1,6,5) -120.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Log file
https://wiki.ch.ic.ac.uk/wiki/index.php?title=File:AJ3318_PF5_OPTF_POP.LOG
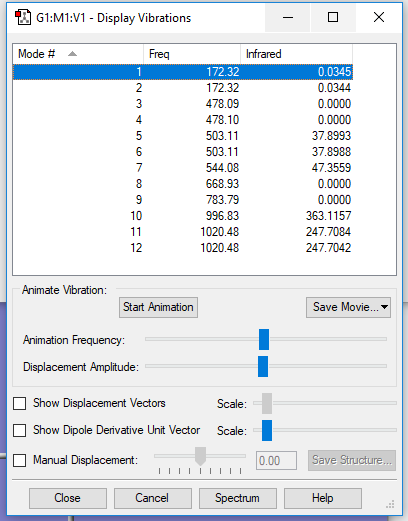
Interactive image
H2 molecule |
Display Vibrations window
Table of vibrational modes
| wavenumber/cm-1 | 172 | 172 | 478 | 478 | 503 | 503 |
| symmetry | E' | E' | E" | E" | E' | E' |
| intensity/arb. u. | 0 | 0 | 0 | 0 | 38 | 38 |
| image |  |
 |
 |
 |
 |
|
| wavenumber/cm-1 | 544 | 669 | 784 | 997 | 1020 | 1020 |
| symmetry | A2" | A1' | A1' | A2" | E' | E' |
| intensity/arb. u. | 47 | 0 | 0 | 363 | 248 | 248 |
| image |  |
 |
 |
 |
 |
|
Charge distribution NBO
Information on MO
General information: 94 MOs output, assume that z axis is directed from the central P atom to an axial F atom.
Marking
Note: All grades and comments are provisional and subjecct to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have recieved your grade from blackboard.
Wiki structure and presentation 1/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES - however, a main part of the wiki are the links in the table of contents which are created by the use of different sections.
Do you effectively use tables, figures and subheadings to communicate your work?
YES
NH3 0.5/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES, however not only mode 1 is highly symmetric but mode 4 is as well. You missed to explain the charges on the atoms of NH3 by an electronegativity argument
N2 and H2 0.5/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES, you could have explained that the charges are 0 as the electronegativities are equal.
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES
Haber-Bosch reaction energy calculation 1/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 3/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
You have done a good job of presenting this information, well done! You should have commented on the calculated vibration modes (degeneracy, which ones should be visible in an experimental spectrum...) You should have explained the charges using an electronegativity argument. You correctly identified the AOs contributing to the MOs correctly in most cases and gave their energies. For the first two MOs you correctly described the bonding situation. From the third MO on you are confused about the bonding situation. You cannot describe only one part of the MO as bonding and another part as anti-bonding (e.g. the third MO is a bonding one.) For the description of the last 2 MOs cancelling out electron density is not the reason why not all AOs participate in forming the MO!
Independence 1/1
If you have finished everything else and have spare time in the lab you could: Check one of your results against the literature, or
YES - well done! You checked not only one but both - N2 and H2 - against information from crystal structures.
Do an extra calculation on another small molecule, or Do some deeper analysis on your results so far