Rep:Mod:aec18
NH3 Ammonia Molecule
Optimised molecule information
| Calculation method | RB3LYP |
| Basis set | 6-31G(d.p) |
| Final energy (RB3LYP) | -56.5577687 a.u. |
| Point group | C3v |
| RMS gradient | 0.00000485 |
| Bond distance | 1.02Å |
| Bond angle | 106° |
The optimisation file is liked to here
Item Table
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986282D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7412 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7412 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7412 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8571 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Jmol dynamic image
Ammonia |
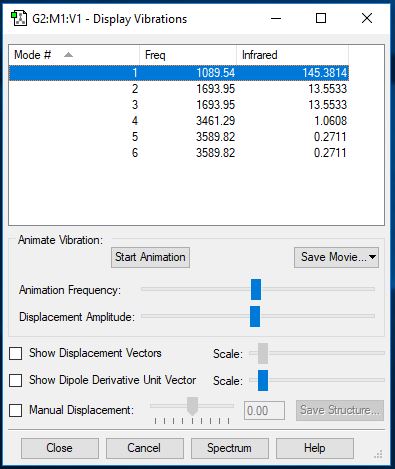
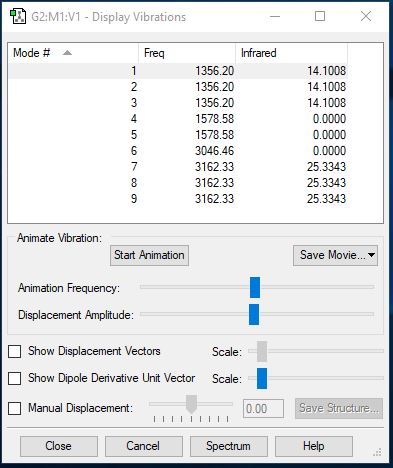
NH3 Vibrations
| Wavenumber (cm-1) | 1090 | 1693 | 1693 | 3461 | 3590 | 3590 |
| Symmetry | A1 | E | E | A1 | E | E |
| Intensity (arbitrary units) | 145 | 13.6 | 13.6 | 1.06 | 0.27 | 0.27 |
| Image |  |
 |
 |
 |
 |
|
Questions
How many modes do you expect from the 3N-6 rule?
6 modes of vibration can be expected from this molecule.
Which modes are degenerate (ie have the same energy)?
The two vibrations of wavelength 1693 and 3590 are degenerate.
Which modes are "bending" vibrations and which are "bond stretch" vibrations?
Bending = 1089cm-1, 1693cm-1, 1693cm-1 Bond stretch = 3461cm-1, 3589cm-1, 3589cm-1
Which mode is highly symmetric?
1089cm-1 and 3461cm-1 modes are highly symmetric.
One mode is known as the "umbrella" mode, which one is this?
1089cm-1 is the 'umbrella' mode.
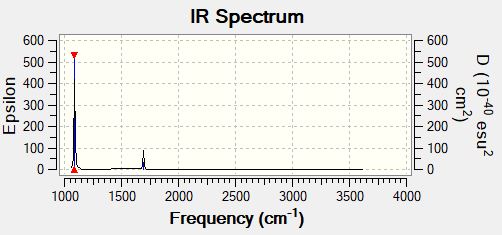
How many bands would you expect to see in an experimental spectrum of gaseous ammonia?
2 bands can be expected:
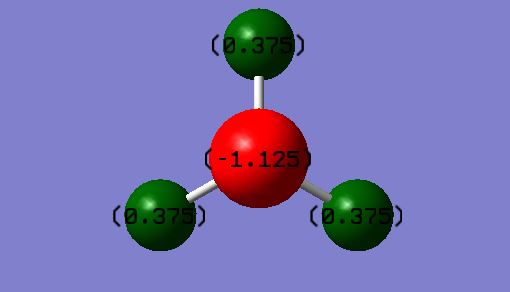
Atomic charges
As shown on image:
H atoms = 0.375
N atom = -1.125
This is as expected as the nitrogen atom is the more electronegative of the two and so will carry the negative charge.
N2 Nitrogen Molecule
Optimisation information
| Calculation method | RB3LYP |
| Basis set | 6-31G(d.p) |
| Final energy (RB3LYP) | -109.5241287 a.u. |
| Point group | DinfH |
| RMS gradient | 0.00000060 |
| Bond distance | 1.11Å |
The optimisation file is liked to here
Item table
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.400954D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1055 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Jmol dynamic image
Nitrogen |
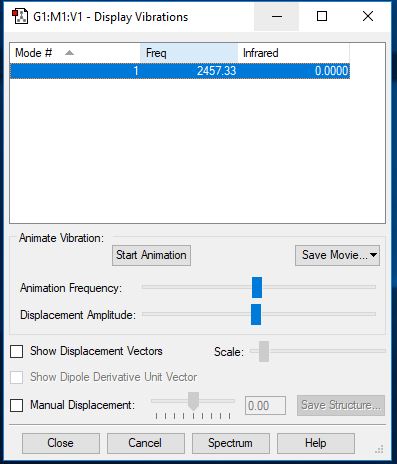
N2 Vibrations
| Wavenumber (cm-1) | 2457 |
| Symmetry | SGG |
| Intensity (arbitrary units) | 0.00 |
| Image | 
|
Atomic Charges
H2 Hydrogen Molecule
Optimisation information
| Calculation method | RB3LYP |
| Basis set | 6-31G(d.p) |
| Final energy (RB3LYP) | -1.1785394 a.u. |
| Point group | DinfH |
| RMS gradient | 0.00000017 |
| Bond distance | 0.74Å |
The optimisation file is liked to here
Item table
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 0.7428 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Jmol dynamic image
Hydrogen molecule |
H2 vibrations
| Wavenumber (cm-1) | 4466 |
| Symmetry | SGG |
| Intensity (arbitrary units) | 0.000 |
| Image | 
|
Atomic Charges
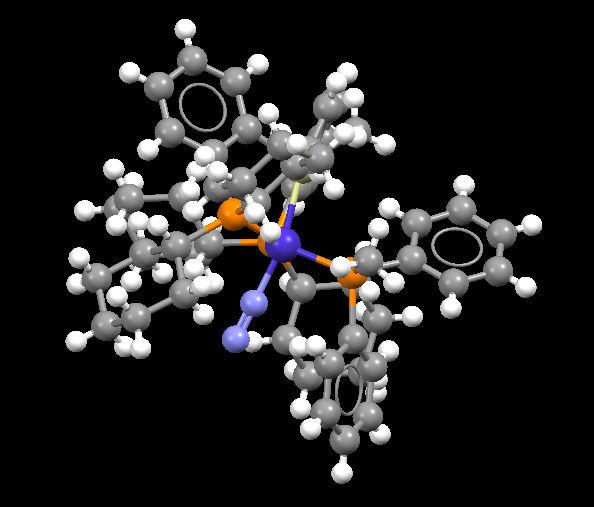
Transition metal complex (containing N2)
The mono-metallic TM complex containing N2 has the code VEJSOV. It can be found here: [1]
The TM Complex N-N bond length = 1.1165 Å
Optimised N2 molecule bond length = 1.11 Å
The N-N bond length in the TM complex is slightly longer than the optimised N-N bond length for N2 and so can therefore be considered as slightly weaker. This is because for N2 the electrons are uniformly shared within the triple bond, making it stronger as there is little electron distortion in the bond between the two atoms. For the TM complex, one of the nitrogen atoms is also bonded to a Co atom and so the electron distribution within the N-N bond is less as some of these electrons are now distorted towards the N-Co bond. This therefore makes the N-N bond weaker and so longer which explains the observations above.
Although the value worked out for the N-N bond length in my N2 optimised molecule is similar to the experimental value calculated for the TM complex using Mercury, errors must be taken into account. These include both computational and experimental errors. To decrease computational errors a better method could have been used, such as CCSD instead of B3LYP which is what I used.
Transition metal complex (containing H2)
The mono-metallic TM complex containing H2 has the code CEJDEA. It can be found here: [2]
The TM Complex N-N bond length = 0.7552 Å
Optimised H2 molecule bond length = 0.74Å
As previously suggested, the increased bond length in the TM complex is due to the other atom bonded to the two hydrogen's (tungsten in this case), resulting in the lengthening of the H-H bond as electron density in the bond is less. There is a greater effect for the lengthening of the hydrogen bond than the nitrogen bond (see previous) as the H-H bond is a single bond rather than a triple bond and so the effect caused by another atom on this bond is greater.
Haber process calculations
The energy required for the reaction Nx2 + 3Hx2 -> 2NHx3 can be determined using the final energy values previously calculated.
(Energies quoted are in a.u. unless otherwise specified).
E(NH3)=-56.5577687
2*E(NH3)=-113.1155374
E(N2)= -109.5241286
E(H2)= -1.17853936
3*E(H2)=-3.53561808
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -113.1155374 - [-109.5241286 + (-3.53561808)] ΔE= -113.1155374 + 113.05974668 = -0.05579072
ΔE= -0.05579 a.u. = -146.5 kJ/mol
The energy for converting hydrogen and nitrogen into ammonia is -146.5 kJ/mol. The combined energies of the reactants is higher (less negative) than the energy of the ammonia product and so the ammonia product is the more stable. The reaction is exothermic which further supports this theory.
Molecule of my choice: CH4
Optimisation information
| Calculation method | RB3LYP |
| Basis set | 6-31G(d.p) |
| Final energy (RB3LYP) | -40.5240140 a.u. |
| RMS gradient | 0.0000320 |
| C-H Bond distance | 1.09Å |
| Bond angle | 109° |
The optimisation file is liked to here
Item table
Item Value Threshold Converged?
Maximum Force 0.000063 0.000450 YES
RMS Force 0.000034 0.000300 YES
Maximum Displacement 0.000179 0.001800 YES
RMS Displacement 0.000095 0.001200 YES
Predicted change in Energy=-2.256038D-08
Optimization completed.
-- Stationary point found. ----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.092 -DE/DX = -0.0001 !
! R2 R(1,3) 1.092 -DE/DX = -0.0001 !
! R3 R(1,4) 1.092 -DE/DX = -0.0001 !
! R4 R(1,5) 1.092 -DE/DX = -0.0001 !
! A1 A(2,1,3) 109.4712 -DE/DX = 0.0 !
! A2 A(2,1,4) 109.4712 -DE/DX = 0.0 !
! A3 A(2,1,5) 109.4712 -DE/DX = 0.0 !
! A4 A(3,1,4) 109.4712 -DE/DX = 0.0 !
! A5 A(3,1,5) 109.4712 -DE/DX = 0.0 !
! A6 A(4,1,5) 109.4712 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -120.0 -DE/DX = 0.0 !
! D2 D(2,1,5,3) 120.0 -DE/DX = 0.0 !
! D3 D(2,1,5,4) -120.0 -DE/DX = 0.0 !
! D4 D(3,1,5,4) 120.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Jmol dynamic image
Methane |
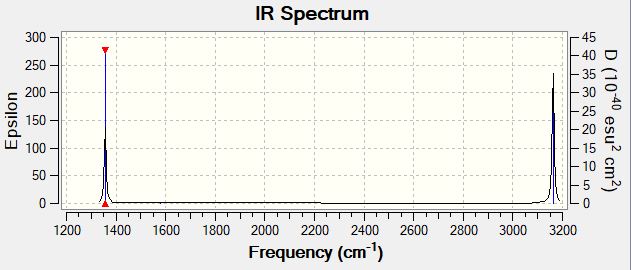
CH4 Vibrations
| Wavenumber (cm-1) | 1356 | 1356 | 1356 | 1579 | 1579 | 3046 | 3162 | 3162 | 3162 |
| Symmetry | T2 | T2 | T2 | E | E | A1 | T2 | T2 | T2 |
| Intensity (arbitrary units) | 14.10 | 14.10 | 14.10 | 0.00 | 0.00 | 0.00 | 25.33 | 25.33 | 25.33 |
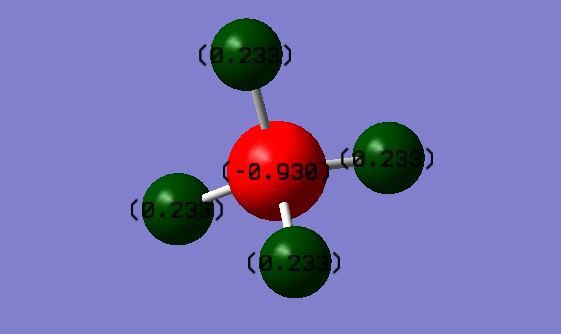
Atomic charges
H atom charge = 0.233
C atom charge = -0.930
This is as expected as the carbon atom is the more electronegative of the two and therefore will carry the negative charge.
Molecular Orbitals
Marking
Note: All grades and comments are provisional and subject to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have received your grade from blackboard.
Wiki structure and presentation 0.5/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES, overall a well structured wiki, however a lot of your images are oversized preventing the reader from viewing the figures in context as they fill so much of the screen!
NH3 1/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES
N2 and H2 0.5/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES, well done for good explanations and looking up a crystal structure for both molecules.
Haber-Bosch reaction energy calculation 1/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 3.5/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
YES, overall you have some good explanations on the charges and some of the MOs, especially MOs 1 and 2. For MO 3 and 4 you could have mentioned that these MOs increase the bonding between the H atoms and the C atom. MO 6 is actually the antibonding counterpart to MO2, it has the same atomic contributions, but the phase of the C 2s AO is reversed. The H atoms primarily contribute 1s AOs not 2s AOs to the LUMO.
Independence 1/1
If you have finished everything else and have spare time in the lab you could:
Check one of your results against the literature, or
Do an extra calculation on another small molecule, or
Do some deeper analysis on your results so far
You looked up two crystals well done!