Rep:Mod:ac4610
Module 2: Inorganic Modelling
In this lab quantum chemistry is used to look at structures and bonding. Gaussian allows us to predict properties of different molecules, for example geometry, bond lengths, bond angles and IR spectrum. One of the other advantages of Gaussian is that you can analyse molecules that in reality would be difficult to analyse due to toxicity or instability. DFT calculations with the B3LYP function are used for all the calculations.
Week 1
Analysing an optimised BH3
In this optimisation a basis set of 3-21G is used. This has a very low accuracy but it means the calculation runs a lot faster. By running an optimisation we are determining the optimium position of the nuclei for a given electronic configuration.
File type: log file, Link: File:BH3 OPT.LOG
B-H bond length is: 1.19 Å
H-B-H angle is: 120.0°
Calculation type: Optimisation
Calculation Method: B3LYP
Basis Set: 3-21G
Final energy:-26.4623 a.u.
Gradient:0.0002 a.u.
Dipole Moment: 0.00
Point group: D3H
Length of Calculation: 19.0 seconds
Item Value Threshold Converged?
Maximum Force 0.000413 0.000450 YES
RMS Force 0.000271 0.000300 YES
Maximum Displacement 0.001610 0.001800 YES
RMS Displacement 0.001054 0.001200 YES
Predicted change in Energy=-1.071764D-06
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1935 -DE/DX = 0.0004 !
! R2 R(1,3) 1.1935 -DE/DX = 0.0004 !
! R3 R(1,4) 1.1935 -DE/DX = 0.0004 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
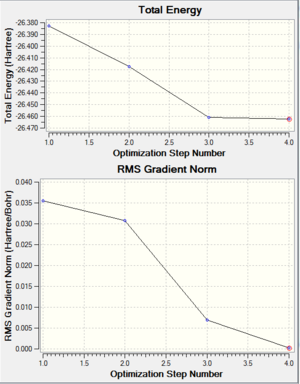
The table shows that the calculation has converged therefore confirming that the structure has been optimised. Another way to confirm the structure has been optimised is to compare the bond length of 1.19 Å to the literature value of 1.19 Å[1]. Since these are the same we can confirm that the structure is optimised. (See below for the 6-31G optimisation graphs to also confirm an optimisation has been achieved)
6-31G(d,p) BH3 Optimisation
In this optimisation 6-31G(d,p) is used as the basis set this is more accurate than the 3-21G so the calculation takes longer to run.
File type: log file, Link: File:BH3 OPT 2.LOG
B-H bond length is: 1.19 Å
H-B-H angle is: 120.0°
Calculation type: Optimisation
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final energy: -26.6153 a.u.
Gradient: 0.0000 a.u.
Dipole Moment: 0.00
Point group: D3H
Length of Calculation: 8.0 seconds
Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.000019 0.001800 YES
RMS Displacement 0.000012 0.001200 YES
Predicted change in Energy=-1.304899D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1923 -DE/DX = 0.0 !
! R2 R(1,3) 1.1923 -DE/DX = 0.0 !
! R3 R(1,4) 1.1923 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
The table shows that the calculation has converged therefore confirming that the structure has been optimised. The graph also shows the gradient approaching zero and therefore showing the structure has been optimised. When looking at the animation of how the structure varies in relation to the graph, the first image has no bonds. This does not necessarily mean there are no bonds because gaussview draws bonds depending on the distance. In reality a bond is a high electron density between atoms which causes an attraction so a mere distance relationship is insufficient to describe a bond. This is a limitation of gaussview since just because it doesn't show a bond doesn't mean it is not there. The bond length for this is the same as calculated with the weaker basis set and is therefore also consistent with the literature value of 1.19Å.[2]
TlBr3 Optimisation HPC
In this optimisation pseudo-potentials are used since thalium and bromine are heavier atoms and therefore normal calculations would take too long. Pseudo-potentials can only be used on heavy atoms since the assumption that the valence electrons dominate in bonding interactions holds well.
The link to my log file is File:TlBr3 optHPC.log
D-space link: DOI:10042/22457
Tl-Br bond length is: 2.65 Å
Br-Tl-Br angle is: 120.0°
Calculation type: FOPT
Calculation Method: RB3LYP
Basis Set: LANL2DZ
Final energy:-91.2181 a.u.
Gradient:0.0000 a.u.
Dipole Moment: 0.00
Point group: D3H
Job Calculation time: 24.4 s
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000014 0.001200 YES
Predicted change in Energy=-6.083977D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.651 -DE/DX = 0.0 !
! R2 R(1,3) 2.651 -DE/DX = 0.0 !
! R3 R(1,4) 2.651 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
The table shows that the calculation has converged therefore confirming that the structure has been optimised. To support this a comparison to literature needs to be made. The bond length measured was 2.65 Å and the angle measured was 120.0º. In literature the experimental value is measured to be 2.512Å.[3]
BBr3 Optimisation HPC
In this optimisation a full basis set was used on the B but PPs on the Br atoms since PP assumptions are inaccurate for lighter atoms such as boron.
The link to my log file is File:BBr3 optHPC.log
Link to D-space is DOI:10042/22455
B-Br bond length is: 2.02 Å
Br-B-Br angle is: 120.0°
Calculation type: FOPT
Calculation Method: RB3LYP
Basis Set: Gen
Final energy:-64.4290 a.u.
Gradient:0.0142 a.u.
Dipole Moment: 0.00
Point group: D3H
Job Calculation time: 22.0 s
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000036 0.001800 YES
RMS Displacement 0.000023 0.001200 YES
Predicted change in Energy=-4.027553D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.934 -DE/DX = 0.0 !
! R2 R(1,3) 1.934 -DE/DX = 0.0 !
! R3 R(1,4) 1.934 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
From the table we can see the structure has optimised and the bond length calculated is 2.02Å this is greater than the literature value of 1.893Å.[4] The difference in bond length is due to us using PPs on the Br atom. By using PPs on the bromine atoms we are assuming the non-valence orbitals don't affect the bonding however we know this is not true and hence the difference in the literature value and the calculated value.
Comparison of Bond lengths
| Molecule | Bond | Length (a.u.) |
|---|---|---|
| BH3 | B-H | 1.19 |
| TlBr3 | Tl-Br | 2.65 |
| BBr3 | B-Br | 2.02 |
The B-H bond is shorter than the B-Br bond which means it is also stronger. This is because the orbital overlap is better between boron and hydrogen as the orbitals are a similar size, bromine is a much bigger atom so there is an orbital size mismatch hence weaker over lap and a weaker, longer bond. The similarity between hydrogen and bromine is that they both contribute 1 electron to a bond. The difference between them is the size difference discussed above.
The B-Br bond is shorter and therefore stronger than the Tl-Br. This is once again due to the size of the atoms. Tl and B are both in group 3 of the periodic table so both bond in the same way however Tl is much bigger than B and therefore has much more diffuse orbitals. This means any bonds it forms will be weaker and hence longer.
Frequency analysis for BH3
The link to my log file is here
B-H bond length is: 1.19 Å
H-B-H angle is: 120.0°
Calculation type: Frequency
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final energy:-26.6153 a.u.
Gradient:0.0000 a.u.
Dipole Moment: 0.0
Point group: D3H
Length of Calculation: 6.0 seconds
Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000002 0.000300 YES
Maximum Displacement 0.000019 0.001800 YES
RMS Displacement 0.000009 0.001200 YES
Predicted change in Energy=-1.323374D-10
Optimization completed.
-- Stationary point found.
Frequencies obtained from real output file
Low frequencies --- -0.9033 -0.7343 -0.0054 6.7375 12.2491 12.2824
Low frequencies --- 1163.0003 1213.1853 1213.1880
The file has converged properly and all the low frequencies are within 15 and -15 and all the frequencies recorded are positive so we can conclude that a minima has been found.
| Motion | Description | Frequency | Intensity | Symmetry D3h point group |
|---|---|---|---|---|
| Out of plane wagging | The hydrogen atoms move in and out of the plane and the boron atom moves in the opposite direction at all times. | 1163 | 93 | A2" |
| Scissoring | Two hydrogens atoms move down towards each other and then away from each other in the same plane. When the hydrogens move down the Boron and remaining hydrogen counter act this by moving up. | 1213 | 14 | E' |
| In plane wagging | Two of the hydrogens move, with no change in the angle between them, the other hydrogen moves from side to side in the same plane. The boron atom moves slightly to counter act these movements. | 1213 | 14 | E' |
| Symmetric Stretching | The boron atom stays still and all the hydrogens move away from the boron at the same time in the same plane. | 2582 | 0.00 | A1' |
| Asymmetric stretching | Two of the hydrogen bond angles stretch and contract in alternating fashion. The boron and remaining hydrogen move slightly from side to side to counter act this movement. | 2715 | 126 | E' |
| Asymmetric stretching | Two of the hydrogen-boron bonds stretch and contract in unison while the remaining hydrogen-boron bond stretches in contrast to the other two. The boron atom moves up and down to counter act these movements. | 2715 | 126 | E' |
Image to show the different movements described above (by right clicking on the image you can see the different vibrations):
Vibration |
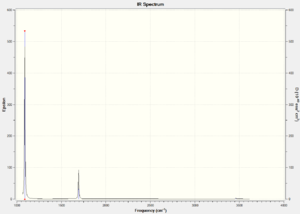
Ir spectra of BH3

There are only 3 peaks because the symmetric stretch will not show up also the 2nd and 3rd stretches are degenerate as are the 5th and 6th.
Frequency analysis for TlBr3
The link to the d-space is DOI:10042/22458
The link to my log file is here
B-H bond length is: 2.65 Å
H-B-H angle is: 120.0°
Calculation type: Frequency
Calculation Method: RB3LYP
Basis Set: LANL2DZ
Final energy:-91.2181 a.u.
Gradient:0.0000 a.u.
Dipole Moment: 0.0
Point group: D3H
Length of Calculation: 16.3 seconds
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000011 0.001200 YES
Predicted change in Energy=-5.660901D-11
Optimization completed.
-- Stationary point found.
Low frequencies --- -3.4213 -0.0026 -0.0004 0.0015 3.9367 3.9367
Low frequencies --- 46.4289 46.4292 52.1449
The file has converged properly and all the low frequencies are within 15 and -15 and all the frequencies recorded are positive so we can conclude that a minima has been found. The lowest real normal mode is 46cm-1.

| TlBr3 | BH3 | ||||
|---|---|---|---|---|---|
| Frequency | Intensity | Symmetry | Frequency | Intensity | Symmetry |
| 46 | 4 | E' | 1163 | 93 | A2" |
| 46 | 4 | E' | 1213 | 14 | E' |
| 52 | 6 | A2" | 1213 | 14.0589 | E' |
| 165 | 0 | A1' | 2582 | 0.0000 | A1' |
| 211 | 25 | E' | 2715 | 126.3307 | E' |
| 211 | 25 | E' | 2715 | 126.3211 | E' |
There is a large difference in the vibrations for TlBr3 and BH3 because the bonds in TlBr3 are longer and weaker and therefore vibrate at higher frequencies. This is because Tl and Br are heavier atoms and so have more diffuse orbitals and the vibrations are slower. There has also been a re-ordering of modes, in TlBr3 the A2" vibration is at a higher frequency than the 2 first E' vibrations whereas in BH3 the A2" vibration has the lowest frequency.
The IR spectra of the TlBr3 and BH3 are similar in that they both have 2 peaks close together at a low frequency and then another single peak at a much higher frequency. The relative intensities of the peaks are also similar since the high frequency peak in both has the highest intensity and the middle vibrational peak has the lowest intensity. There is a large split in the lower 2 vibrations and the higher 2 vibrations. This is because the lower vibrations correspond to translational vibrations and these only result in a small change in dipole whereas the higher vibrations correspond to stretching frequencies that result in a much greater change in dipole. This large change in dipole requires more energy to occur and therefore has a higher frequency.
These frequencies can be compared because the calculations were run using the same basis set. It is important to make sure any comparisons are made only on calculations that have been run using the same basis set because each basis set has a different way of calculating the energies and frequencies. Each basis set starts from a different pint on the potential curve and so when it converges will end at a different energy even if it is run on the same molecule.
We carry out a frequency analysis on molecules to ensure that we have in fact reached a minima instead of a transition state. Since the frequencies calculations are a second derivative if all the frequencies are positive we can conclude that it is a minima. The low frequencies are the frequencies related to vibrational and translational modes and these should be zero however due to the approximations made by the basis set used these are usually between -15 and 15.
Molecular Orbitals of BH3
The link to the d-space is DOI:10042/22461
The link to my fch file is here
B-H bond length is: 2.65 Å
H-B-H angle is: 120.0°
Calculation type: SP
Calculation Method: RB3LYP
Basis Set: 6-31G(D,P)
Final energy:-26.6153 a.u.
Gradient:0.0000 a.u.
Dipole Moment: 0.0
Point group: D3H
Length of Calculation: 7 s
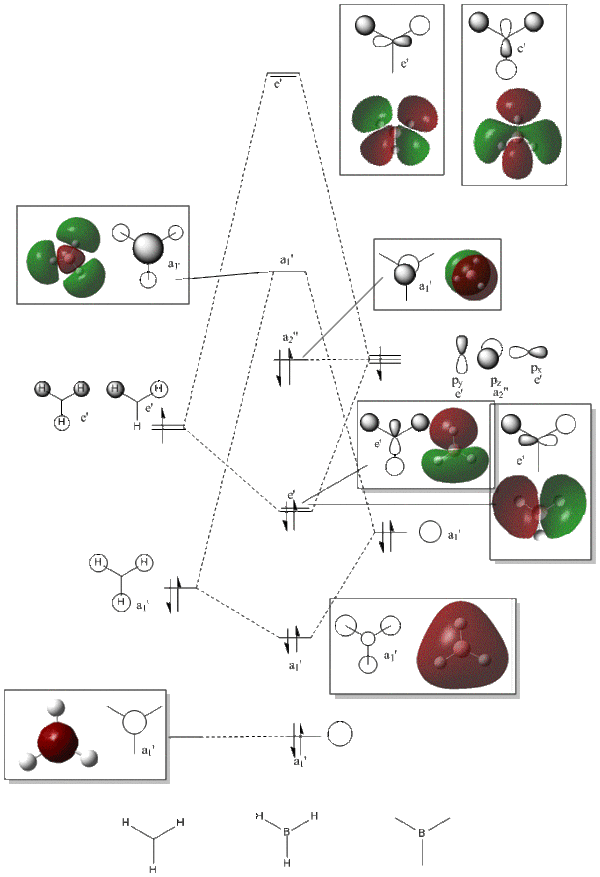
As is shown above the LCAO MOs and the real MOs that were calculated are very similar. The only differences are in how diffuse the real MOs are and this is mainly due to the way we present LCAOs. Therefore from this it can be concluded that LCAO analysis and MO theory is relatively accurate with respect to the more simple MOs for molecules studied in this lab. If we then had to represent more complicated anti-bonding orbitals with LCAOs there would be more of a difference between what we could represent and the actual MOs mainly due to 3D visualisation and interactions that are very difficult to represent in 2D diagrams.
NH3 Optimisation, Frequency and Population Analysis
Optimisation
The link to my log file is here
N-H bond length: 1.02Å
H-N-H bond angle: 105.7°
Calculation Type: FOPT
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final Energy: -56.5578 a.u.
RMS Gradient Norm: 0.0000 a.u.
Dipole Moment: 1.85
Point Group: C1
Length of Calculation: 13.0s
Item Value Threshold Converged?
Maximum Force 0.000024 0.000450 YES
RMS Force 0.000012 0.000300 YES
Maximum Displacement 0.000079 0.001800 YES
RMS Displacement 0.000053 0.001200 YES
Predicted change in Energy=-1.629728D-09
Optimization completed.
-- Stationary point found.
Frequency Anlayisis
The link to my log file is here
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
E(RB3LYP): -56.5578
RMS Gradient Norm: 0.0000
Dipole Moment: 1.85
Point Group: C1
Item Value Threshold Converged?
Maximum Force 0.000023 0.000450 YES
RMS Force 0.000011 0.000300 YES
Maximum Displacement 0.000075 0.001800 YES
RMS Displacement 0.000051 0.001200 YES
Predicted change in Energy=-1.476848D-09
Optimization completed.
-- Stationary point found.
Low frequency lines:
Low frequencies --- -10.0854, -0.0012, 0.0014, 0.0016, 9.2235, 14.6778
Low frequencies --- 1089.3146, 1693.9161, 1693.9488
| Frequency | Intensity |
|---|---|
| 1089 | 145 |
| 1694 | 14 |
| 1694 | 14 |
| 3461 | 1 |
| 3590 | 0 |
| 3590 | 0 |
The file has converged properly and all the low frequencies are within 15 and -15 and all the frequencies recorded are positive so we can conclude that a minima has been found.
Population analysis
The link to the chk file is here
Calculation Type: SP
Calculation Method: RB3LYP
Basis Set: 6-31G(D,P)
Total Energy: -56.5578
RMS Gradient Norm: 0.0000
Dipole Moment: 1.85
| MOs | Symmetry |
|---|---|
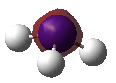 |
E' |
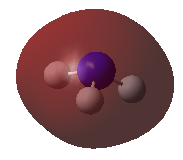 |
E' |
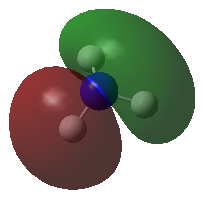 |
A2" |
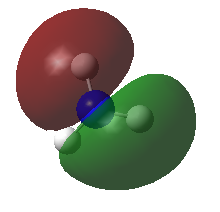 |
A1 |
 |
E1' |
 |
E1' |
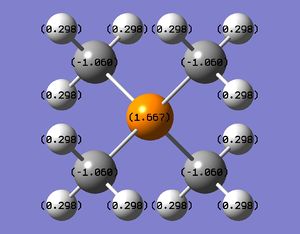
NBO Analysis of NH3
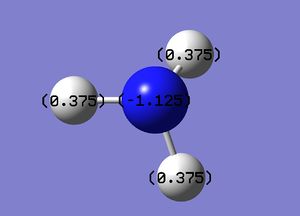
Specific NBO charges of the atoms:
H: 0.375
B: -1.125
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
N 1 -1.12515 1.99982 6.11104 0.01429 8.12515
H 2 0.37505 0.00000 0.62250 0.00246 0.62495
H 3 0.37505 0.00000 0.62250 0.00246 0.62495
H 4 0.37505 0.00000 0.62249 0.00246 0.62495
=======================================================================
* Total * 0.00000 1.99982 7.97852 0.02166 10.00000
Bond composition
 | ||
|---|---|---|
| Bond | Composition | s, p contributions |
| N-H | N: 68.83%, H: 31.17% | N: 24.87% s and 75.05% p, H: 99.91% s |
| N lone pair | N/A | 25.38% s and 74.52% p |
NH3BH3
Optimisation
File type: log, Link: here
Calculation Type: FOPT
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
E(RB3LYP): -83.2247
RMS Gradient Norm: 0.0000
Dipole Moment: 5.57
Point Group: C1
Calculation time: 51.0s
Item Value Threshold Converged?
Maximum Force 0.000124 0.000450 YES
RMS Force 0.000057 0.000300 YES
Maximum Displacement 0.000660 0.001800 YES
RMS Displacement 0.000304 0.001200 YES
Predicted change in Energy=-1.649863D-07
Optimization completed.
-- Stationary point found.
Frequency Analysis
File Type: log, Link: here
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
E(RB3LYP): -83.2247
RMS Gradient Norm: 0.0000
Dipole Moment: 5.56
Point Group: C1
Calculation time: 1min 26.0s
Low frequencies: -5.4652 -2.9941 -0.0011 -0.0007 -0.0005 0.5112
Low frequencies: 263.3728 632.9636 638.4163
| Frequency | Intensity |
|---|---|
| 263 | 0 |
| 633 | 14 |
| 638 | 4 |
| 638 | 4 |
| 1069 | 41 |
| 1069 | 41 |
| 1196 | 109 |
| 1204 | 3 |
| 1204 | 3 |
| 1329 | 114 |
| 1676 | 28 |
| 1676 | 28 |
| 2472 | 67 |
| 2532 | 231 |
| 2532 | 231 |
| 3464 | 3 |
| 3581 | 28 |
| 3581 | 28 |
The file has converged properly and all the low frequencies are within 15 and -15 and all the frequencies recorded are positive so we can conclude that a minima has been found.
Energy Comparison
| Molecule | Energy (a.u.) |
|---|---|
| NH3 | -56.5578 |
| BH3 | -26.6153 |
| NH3BH3 | -83.2247 |
ΔE=E(NH3BH3)-[E(NH3)+E(BH3)]= -83.2247 -(-56.5578 + -26.6153)= -0.0516 a.u.
-0.0516 a.u. = -135.48 kJ/mol
This energy corresponds to the association energy for combining a molecule of NH3 and a molecule of BH3 this also means that the dissociation energy is equal to 135.48 kJ/mol.
Week 2: Ionic Liquids: Designer Solvents
In this mini project computational chemistry is used to predict the properties of ions that could be used in ionic liquids before they are made since there are so many possible combinations of ions that it is impractical to synthesise them all.
Part 1: Comparison of selected 'onium' cations
First Ion: [N(CH3)4]+
Optimisation
File Type: log, Link: here
N-C bond length: 1.51Å
C-N-C bond angle: 109.5°
D space link: DOI:10042/22552
Calculation Type: FOPT
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Charge: +1
Spin: Singlet
E(RB3LYP): -214.18132690
RMS Gradient Norm: 0.00004804
Dipole Moment: 0.00
Point Group: Td
Calculation time: 1 min 10.0 s
Item Value Threshold Converged?
Maximum Force 0.000072 0.000450 YES
RMS Force 0.000029 0.000300 YES
Maximum Displacement 0.000173 0.001800 YES
RMS Displacement 0.000072 0.001200 YES
Predicted change in Energy=-1.017053D-07
Optimization completed.
-- Stationary point found.
Frequency Analysis
File Type: log, Link:here
D-space link: DOI:10042/22582
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Charge: +1
E(RB3LYP): -214.18128421
RMS Gradient Norm: 0.00000282
Dipole Moment: 0.00
Point Group: Td
Calculation time: 4min 49.1s
Low frequencies --- -0.0010 -0.0010 -0.0008 21.3340 21.3340 21.3340
Low frequencies --- 188.4799 292.6202 292.6202
MO Analysis
File Type: chk, Link: here
Calculation Type: SP
Calculation Method: RB3LYP
Basis Set: 6-31G(D,P)
Charge: +1
Total Energy: -214.18132690
RMS Gradient Norm: 0.00000000
Dipole Moment: 0.00
Calculation time: 11.0s
Link to log file for NBO analysis: here
| MOs | Extent of bonding | Annotated diagram showing interactions occurring |
|---|---|---|
 |
Highly Bonding | 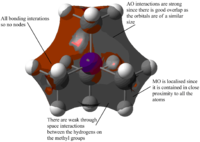 |
 |
Weakly Bonding |  |
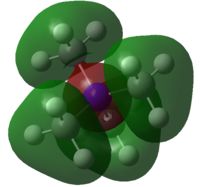 |
Non-Bonding | 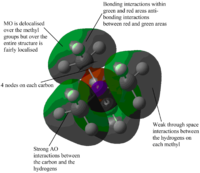 |
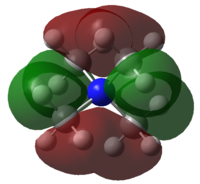 |
Weakly Antibonding | 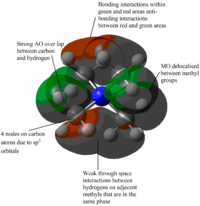 |
 |
Highly Antibonding | 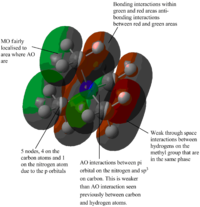 |
Second Ion: [P(CH3)4]+ Optimisation
File Type: log, Link:here
P-C bond length: 1.82Å
C-P-C bond angle: 109.5°
D space link: DOI:10042/22563
Calculation Type: FOPT
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Charge: +1
E(RB3LYP): -500.82717336
RMS Gradient Norm: 0.00003027
Dipole Moment: 0.00
Point Group: Td
Item Value Threshold Converged?
Maximum Force 0.000164 0.000450 YES
RMS Force 0.000038 0.000300 YES
Maximum Displacement 0.000852 0.001800 YES
RMS Displacement 0.000364 0.001200 YES
Predicted change in Energy=-2.442464D-07
Optimization completed.
-- Stationary point found.
Frequency Analysis
File Type: log, Link:here
D space link: DOI:10042/22574
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Charge: +1
E(RB3LYP): -500.82703039
RMS Gradient Norm: 0.00000003
Dipole Moment: 0.00
Point Group: Td
Calculation time: 4min 11.1s
Low frequencies --- -0.0014 0.0028 0.0032 24.7102 24.7102 24.7103
Low frequencies --- 160.0439 194.7830 194.7830
MO analysis
File Type: chk, Link: here
Calculation Type: SP
Calculation Method: RB3LYP
Basis Set: 6-31G(D,P)
Total Energy: -500.82717336
RMS Gradient Norm: 0.00000000
Dipole Moment: 0.00
Calculation time: 9.0s
Link to log file for NBO analysis: here
Third Ion: [S(CH3)3]+
Optimisation
File Type: log, Link:here
S-C bond length: 1.82Å
C-S-C bond angle: 102.8°
D space link: DOI:10042/22576
File Type: log, Link:
Calculation Type: FOPT
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Charge: +1
E(RB3LYP): -517.68327155
RMS Gradient Norm: 0.00004119
Dipole Moment: 0.97
Point Group: C1
Item Value Threshold Converged?
Maximum Force 0.000103 0.000450 YES
RMS Force 0.000035 0.000300 YES
Maximum Displacement 0.001797 0.001800 YES
RMS Displacement 0.000553 0.001200 YES
Predicted change in Energy=-1.242636D-07
Optimization completed.
-- Stationary point found.
Frequency Analysis
File Type: log, Link:here
D-space link: DOI:10042/22634
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Charge: +1
E(RB3LYP): -517.68327146
RMS Gradient Norm: 0.00004118
Dipole Moment: 0.97
Point Group: C1
Calculation time: 3min 36.8s
Low frequencies --- -19.8399 -9.2277 0.0015 0.0033 0.0037 13.7670
Low frequencies --- 161.0240 195.7996 204.5443
MO Analysis
File Type chk, Link:here
Calculation Type: SP
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
E(RB3LYP): -517.68327155 a.u.
RMS Gradient Norm: 0.00000000 a.u.
Dipole Moment: 0.97
Calculation time: 20.0s
Link to log file for NBO analysis: here
Comparison of geometries
| Molecule | Bond length of central atom to methyl group (Å) | Bond Angle (°) | Point group |
|---|---|---|---|
| [N(CH3)3] | 1.51 | 109.5 | Td |
| [P(CH3)3] | 1.82 | 109.5 | Td |
| [S(CH3)3] | 1.82 | 102. 8 | C1 |
[N(CH3)3]+ and [P(CH3)3]+ have a tetrahedral geometry however the bond distance between the central atom and the methyl groups lengthens down the group. This is since P and C have an orbital size mismatch and therefore there is weaker overlap and hence a weaker bond which means it is a longer bond. [S(CH3)3] has the same bond length between the central atom and the methyl groups as [P(CH3)3] since S is in the same row as P the size mismatch between it and the C will be similar to that of P and C. However it does not have a tetrahedral structure since it has a lone pair that causes the bond angle to be smaller than in a tetrahedral due to repulsion of the other bonds with the lone pair.
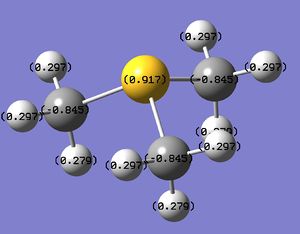
NBO charge and Population Analysis
All three of the ions above have a positive charge, however the charge density of the central atom is different in each case. In the table above we can see that nitrogen is the only central atom that has a negative charge density. This is because when an electron is lost Nitrogen doesn't have a lone pair to lose the electron from so the charge is more delocalised over the whole atom. Therefore the electronegativity difference between nitrogen and carbon becomes the determining factor of the nitrogen charge and since nitrogen has a significantly greater electronegativity than carbon nitrogen has a slightly negative electron density. Sulphur and Phosphorus have a positive charge density this is because both have a lone pair of electrons which is where the electron is most likely to be removed and therefore the positive charge is more localised on the central atom. Phosphorus has a higher charge density that sulphur because it is less electronegative and therefore it more easily loses an electron and also doesn't gain any electron density from the carbon atoms.
From the table above we can see the contributions each of the central atoms make to the C-X bond. When we compare this to the electron distribution calculated before we can see that the atoms that have more negative charge density contribute more to the bond. Therefore nitrogen contributes most out of the 3 central atoms to the C-X bond. This is because a bond is an area of high electron density so if an atom has a low charge density it has more negative charge that it can donate to a bond.
Therefore from these calculations we can see that the traditional description of [N(CH3)4]+ isn't valid since the nitrogen has a more negative electron density and therefore can't have lost an electron. In this case the positive charge is actually located on the hydrogens since these are the only atoms that have a positive charge density in this molecule.
Part 2: Influence of functional groups
[C(CH3)3(CH2CN)]+
Optimisation
File Type: log, Link: here
D-space link: DOI:10042/22605
Calculation Type: FOPT
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Charge: +1
E(RB3LYP): -306.39376934
RMS Gradient Norm: 0.00001767
Dipole Moment: 5.77
Point Group: CS
Calculation time: 6min 15s
Item Value Threshold Converged?
Maximum Force 0.000035 0.000450 YES
RMS Force 0.000010 0.000300 YES
Maximum Displacement 0.000472 0.001800 YES
RMS Displacement 0.000175 0.001200 YES
Predicted change in Energy=-3.185900D-08
Optimization completed.
-- Stationary point found.
Frequency Analysis
File Type: log, Link:here
D-space link: DOI:10042/22604
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Charge: +1
E(RB3LYP): -306.39376217
RMS Gradient Norm: 0.00000193
Dipole Moment: 5.76
Point Group: CS
Calculation time: 15min 44.7s
Low frequencies --- -3.8000 -0.0013 -0.0013 -0.0011 2.6853 3.9802
Low frequencies --- 91.8764 153.9677 212.1260
MO Analysis
File Type: chk, Link: here
Calculation Type: SP
Calculation Method: RB3LYP
Basis Set: 6-31G(D,P)
Total Energy: -306.39376934
RMS Gradient Norm: 0.00000000
Dipole Moment: 5.7651
Calculation time: 41.0s
Link to log file for NBO analysis: here
[C(CH3)3(CH2OH)]+
Optimisation
File Type: log, Link:here
D-space link: DOI:10042/22606
Calculation Type: FOPT
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Charge: +1
E(RB3LYP): -289.39471441
RMS Gradient Norm: 0.00000963
Dipole Moment: 2.13
Point Group: C1
Calculation time: 1hr, 14min, 47.7s
Item Value Threshold Converged?
Maximum Force 0.000018 0.000450 YES
RMS Force 0.000006 0.000300 YES
Maximum Displacement 0.001139 0.001800 YES
RMS Displacement 0.000346 0.001200 YES
Predicted change in Energy=-1.914038D-08
Optimization completed.
-- Stationary point found.
Frequency Analysis
File Type: log, Link:here
D-space link: DOI:10042/22608
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Charge: +1
E(RB3LYP): -289.39470723
RMS Gradient Norm: 0.00000088
Dipole Moment: 2.14
Point Group: C1
Calculation time: 27min 45.6s
Low frequencies --- -8.4062 -5.0188 -1.1556 -0.0008 -0.0007 0.0002
Low frequencies --- 131.1093 213.4637 255.7117
MO Analysis
File Type: chk, Link: here
Calculation Type: SP
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
E(RB3LYP): -289.39471441
RMS Gradient: 0.00000000
Dipole Moment: 2.13
Calculation time: 1min 7.0s
Link to log file for NBO analysis: here
NBO and charge distribution analysis
In the table above we can see the effects -OH and -CN have on the electron distribution of the molecule. The electron density of the Nitrogen becomes more negative when -OH is the ligand. This is because -OH is an electron donating ligand and this therefore increases the electron density on the nitrogen. In comparison the nitrogen electron density in the molecule with a -CN ligand is less negative. This is because the -CN ligand withdraws electron density from the nitrogen.
When we look at the electron distribution for the carbons however this trend isn't apparent. The carbon with the greatest electron density is in the molecule with no ligand attached. This is because even though the -OH ligand is electron donating, oxygen is very electronegative so this causes electron density to be withdrawn from the adjacent carbon making it's electron density positive. The electron distribution for the carbon in the molecule with the -CN ligand is slightly more positive than the carbon atom in [N(CH3)4]+ this is because of the electron withdrawing effect of the -CN ligand.
HOMO AND LUMO COMPARISON
| HOMOs | ||
|---|---|---|
| [N(CH3)4]+ | [C(CH3)3(CH2OH)]+ | [C(CH3)3(CH2CN)]+ |
 |
 |
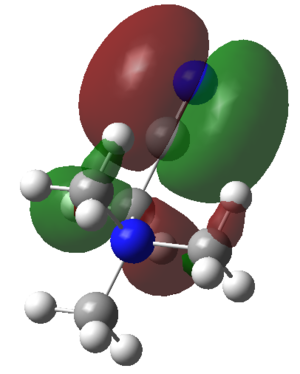 |
| Energy | ||
| -0.57934 | -0.48762 | -0.50045 |
The HOMO of the [N(CH3)4]+ is spread right across the molecule and there are 5 nodes. The HOMO of the [C(CH3)3(CH2OH)]+ is more localised mainly on the -OH and the N atom this is because these are more electronegative. The HOMO of the [C(CH3)3(CH2CN)]+ is very localised to the -CN substituents and the carbon it is connected to this is because of the electron withdrawing effect of the -CN. The HOMO decreased in energy for the [C(CH3)3(CH2CN)]+ ion and then decreased again for the [C(CH3)3(CH2OH)]+ ion.
| HOMOs | ||
|---|---|---|
| [N(CH3)4]+ | [C(CH3)3(CH2OH)]+ | [C(CH3)3(CH2CN)]+ |
 |
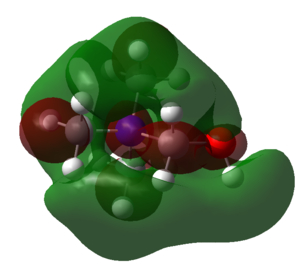 |
 |
| Energy | ||
| -0.13299 | -0.12458 | -0.18179 |
The LUMOs are more diffuse than the HOMOs. For the [N(CH3)4]+ ion the shape of the LUMO like the HOMO is symmetrical and spread evenly across the molecule. In comparison the LUMOs of the molecules with -CN and -OH ligands is much less symmetrical. The energies of the LUMOs has changed, for the [C(CH3)3(CH2OH)]+ ion the energy has decreased whereas for the [C(CH3)3(CH2CN)]+ ion the energy has increased. Overall this means the HOMO-LUMO gaps for [N(CH3)4]+, [C(CH3)3(CH2OH)]+ and [C(CH3)3(CH2CN)]+ respectively are; 0.4464 a.u., 0.3630 a.u. and 0.3187 a.u.. Therefore adding a ligand decreases the HOMO-LUMO band gap, if the ligand is electron withdrawing it decreases it by more than if the ligand is electron donating. The chemical impact of this change is that when the HOMO-LUMO gap is smaller the molecule is more easily reduced.
Conclusion
In conclusion Gaussian has been used effectively to optimise and analyse structure that would otherwise had been dangerous or impractical to investigate in a lab. We saw that changing the atoms in a molecule affects the bond lengths and electron distribution depending on their positions in the periodic table. In the mini project it was demonstrated that changing the central ion has a significant effect on the structure and geometry of the ion. It was also concluded that the traditional view of a [N(CH3)4]+ ion with the positive charge located on the nitrogen isn't correct. In addition when ligands are added to an ion this affects the HOMO-LUMO band gap and the charge distribution. This has an effect on how easily the ion can be reduced.
References
- ↑ W.M Haynes ' 'CRC handbook of Chemistry and Physics' ' 2011 92nd Ed. 9-21
- ↑ W.M Haynes ' 'CRC handbook of Chemistry and Physics' ' 2011 92nd Ed. 9-21
- ↑ J. Glaser and G. Johansson, Acta Chemica Scandinavica 1982, 36a, 125-135 DOI:10.1021/jp0124802
- ↑ W.M Haynes; CRC Handbook of Chemistry and Physics, 2011, 92nd Ed., 9-20