Rep:Mod: WXY2816
NH3 Molecule
Information of NH3 Molecule
| Molecular Name | Ammonia |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Energy E(RB3LYP) in atomic units(au) | -56.55776873 |
| RMS Gradient Norm in atomic units(au) | 0.00000485 |
| Point Group | C3V |
| N-H bond length | 1.01798 Å |
| H-N-H bond angle | 105.741° |
The literature values of N-H bond length is 101.7 pm which is 1.017 Å and H-N-H bond angle is 106.7°[1]. Both calculated values are close to theoretical values. Hence, calculation in GaussView using quantum mechanical methods is very accurate.
Item Table
Item Value Threshold Converged? Maximum Force 0.000066 0.000450 YES RMS Force 0.000066 0.000300 YES Maximum Displacement 0.000087 0.001800 YES RMS Displacement 0.000123 0.001200 YES Predicted change in Energy=-5.726834D-09
Jmol Dynamic Image
NH3 molecule |
The optimisation file is linked to here
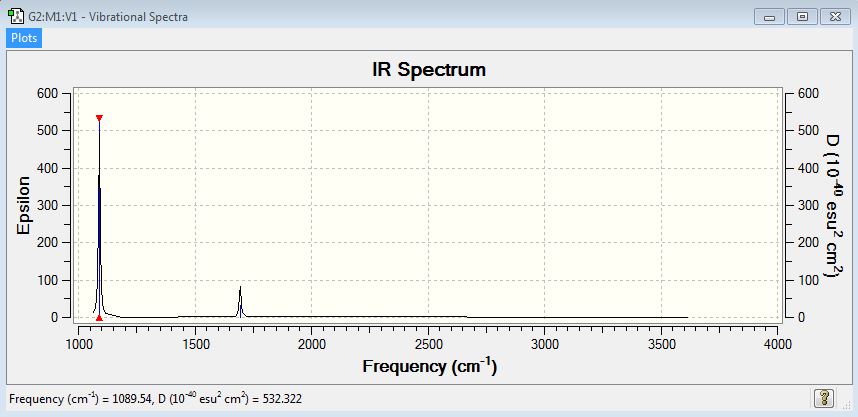
Vibrational Frequencies
NH3 Vibrational Frequencies Table is shown below
| Mode Number | Wavenumber | Vibrational Mode | Relative Intensity |
| 1 | 1089.54 | N-H wagging | 145.3814 |
| 2 | 1693.95 | H-N-H scissoring (bending) | 13.5533 |
| 3 | 1693.95 | H-N-H scissoring (bending) | 13.5533 |
| 4 | 3461.29 | N-H symmetric stretching | 1.0608 |
| 5 | 3589.82 | N-H asymmetric stretching | 0.2711 |
| 6 | 3589.82 | N-H asymmetric stretching | 0.2711 |
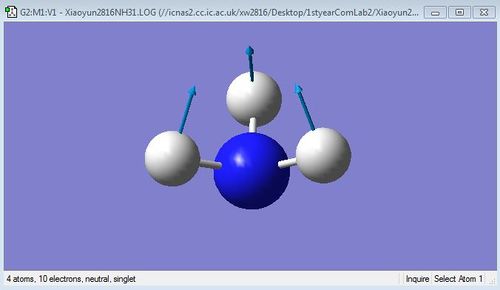
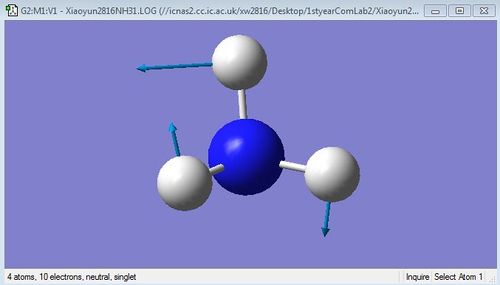
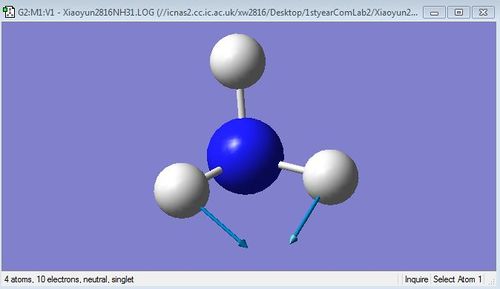
The images below are 6 different vibrational modes of NH3 molecule:
N-H wagging
H-N-H scissoring (bending)
H-N-H scissoring (bending)
N-H symmetric stretching
N-H asymmetric stretching
N-H asymmetric stretching
Questions
1. How many modes do you expect from the 3N-6 rule?
Ammonia is non-linear and follows 3N-6 rule. There are four atoms in one ammonia molecule. 3×4-6=6, six vibrational modes are expected.
2.which modes are degenerate (ie have the same energy)?
Vibrational modes 2 and 3 are degenerate. Vibrational modes 5 and 6 are degenerate.
3.which modes are "bending" vibrations and which are "bond stretch" vibrations?
Vibrational modes 1,2 and 3 (wagging and scissoring modes) are "bending" vibrations. Vibrational modes 4, 5 and 6 (stretching modes) are "bond stretch" vibrations.
4.which mode is highly symmetric?
Vibrational mode 4 is highly symmetric. (symmetric stretching)
5.one mode is known as the "umbrella" mode, which one is this?
Vibrational mode 1 is the "umbrella" mode. (wagging)
6.how many bands would you expect to see in an experimental spectrum of gaseous ammonia?
Two obvious bands are expected to see in an IR spectrum. The two bands are seen at around 1090 cm-1 and 1694 cm-1. The intensities of symmetrical stretching mode and asymmetrical stretching modes are 1.0608 and 0.2711 cm-1. They are too small to be clearly seen in the IR spectrum.
Charges on NH3 molecule
Charge on N atom: -1.125 e-
Charge on H atom: 0.375 e-
what charge (positive or negative) would you expect for N and H and why
Nitrogen atom is more electronegative than hydrogen atom. Thus N atom is expected to have a negative charge while H atom is expected to have a positive charge.
N2 Molecule
Information of N2 Molecule
| Molecular Name | Nitrogen |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Energy E(RB3LYP) in atomic units(au) | -109.52412868 |
| RMS Gradient Norm in atomic units(au) | 0.00000060 |
| Point Group | D∞h |
| Nitrogen-Nitrogen bond length | 1.10550Å |
The literature value of bond length of nitrogen-nitrogen triple bond is 1.0976 Å[2] which is very close to the calculated value. Hence, calculation using quantum mechanical methods is extremely accurate. The small difference may arise from the approximations used in the calculation.
Item Table
Item Value Threshold Converged? Maximum Force 0.000001 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES Predicted change in Energy=-3.401002D-13
Jmol Dynamic Image
N2 molecule |
The optimisation file is linked to here
Vibrational Frequencies
N2 Vibrational Frequencies Table is shown below
N2 molecule is linear and only has one vibrational mode (symmetrical streting) which is at 2457.33 cm-1.
H2 Molecule
Information of H2 Molecule
| Molecular Name | Hydrogen |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Energy E(RB3LYP) in atomic units(au) | -1.17853935 |
| RMS Gradient Norm in atomic units(au) | 0.00003809 |
| Point Group | D∞h |
| H-H bond length | 0.74289Å |
The literature value of H-H bond length is 0.74 Å.[3] Calculated value comply with the literature value. Hence, calculation in GaussView is very accurate.
Item Table
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES Predicted change in Energy=-5.986271D-10
Jmol Dynamic Image
H2 molecule |
The optimisation file is linked to here
Vibrational Frequencies
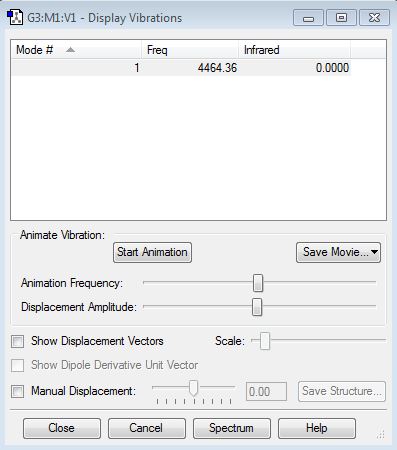
H2 Vibrational Frequencies Table is shown below
H2 molecule is linear and only has one vibrational mode (symmetrical streting) which is at 4464.36 cm-1.
Haber-Bosch reaction energy calculation
Calculations
E(NH3)= -56.55776873 au
2*E(NH3)= -113.11553746 au
E(N2)= -109.52412868 au
E(H2)= -1.17853935 au
3*E(H2)= -3.53561805 au
ΔE=2×E(NH3)-[E(N2)+3×E(H2)]= -113.11553746-(-109.52412868-3.53561805) = -0.05579073 au = -0.05579073×2625.5 kJ/mol = -146.48 kJ/mol (2d.p.)
Since the reaction is exothermic, the product ammonia gas is more stable than gaseous reactants.
The literature value of Haber-Bosch reaction is ΔE = -91.4 kJ/mol[4] The calculated reaction energy deviates from the literature value and the difference is large. This is because there are many assumptions in calculating energies of molecules in GaussView. For example the calculated energy of the molecule in GaussView is not the same as the energy of the molecule in standard state. Hence, the calculated energy of reaction from GaussView is not that accurate.
ClF3 Molecule
Information of ClF3 Molecule
| Molecular Name | Chlorine trifluoride |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Energy E(RB3LYP) in atomic units(au) | -759.46531674 |
| RMS Gradient Norm in atomic units(au) | 0.00009315 |
| Point Group | CS |
| Cl-F bond length | 1.72881Å, 1.65163Å |
| F-Cl-F bond angle | 87.119°, 174.238° |
Item Table
Item Value Threshold Converged? Maximum Force 0.000132 0.000450 YES RMS Force 0.000093 0.000300 YES Maximum Displacement 0.000733 0.001800 YES RMS Displacement 0.000455 0.001200 YES Predicted change in Energy=-1.543576D-07
Jmol Dynamic Image
ClF3 molecule |
The optimisation file is linked to here
Vibrational Frequencies
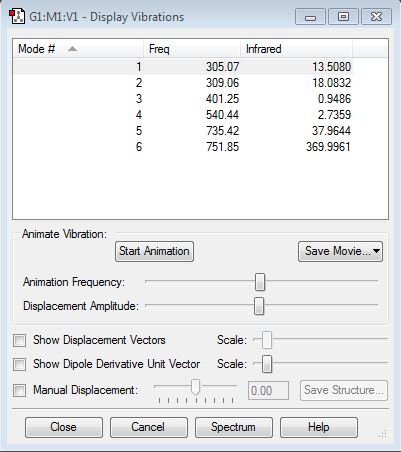
ClF3 Vibrational Frequencies Table is shown below
| Mode Number | Wavenumber | Vibrational Mode | Relative Intensity |
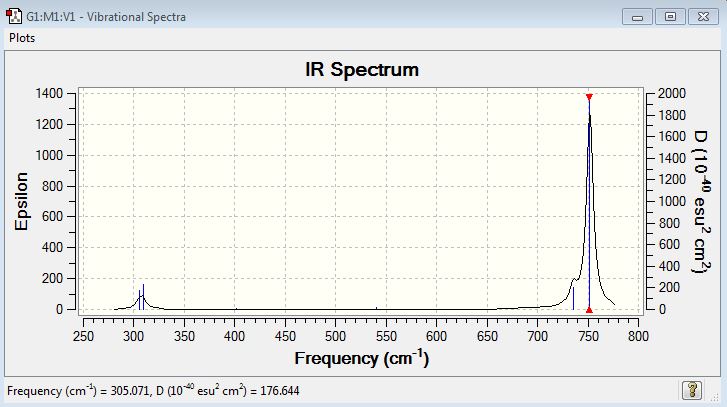
| 1 | 305.07 | F-Cl-F symmetric scissoring (bending) | 13.5080 |
| 2 | 309.06 | F-Cl-F symmetric scissoring (bending) | 18.0832 |
| 3 | 401.25 | F-Cl-F asymmetric scissoring (bending) | 0.9486 |
| 4 | 540.44 | Cl-F symmetric stretching | 2.7359 |
| 5 | 735.42 | Cl-F asymmetric stretching | 37.9644 |
| 6 | 751.85 | Cl-F asymmetric stretching | 369.9961 |
The images below are 6 different vibrational modes of ClF3 molecule:
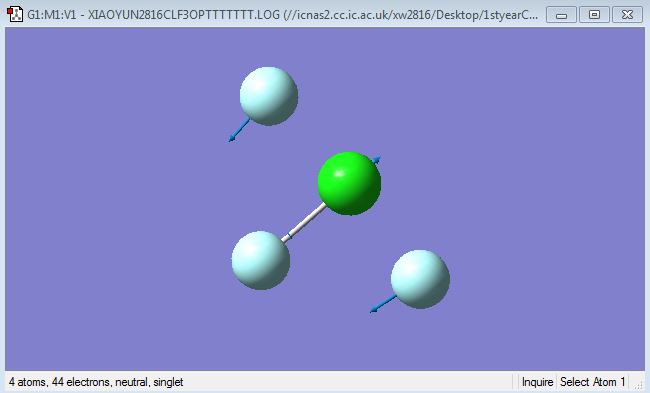
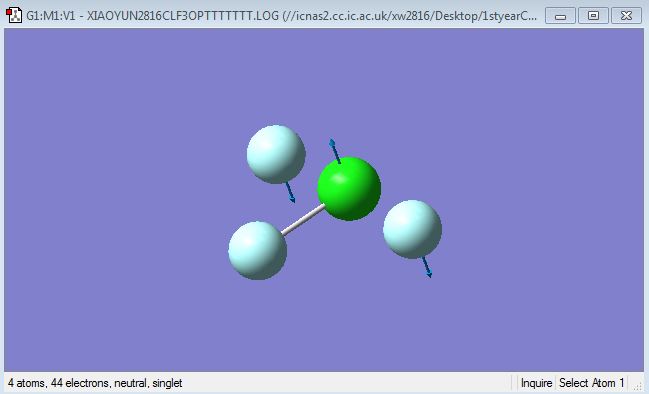
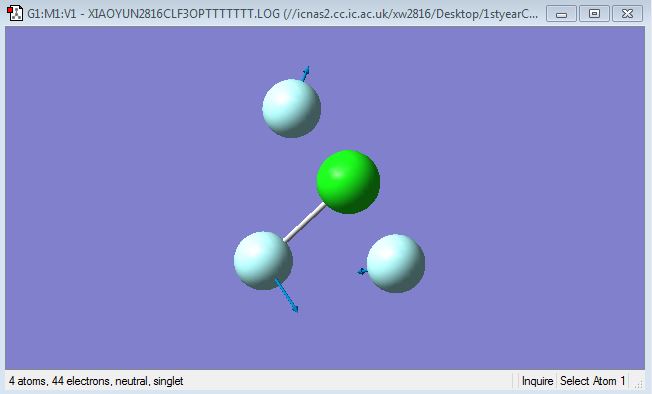
F-Cl-F symmetric scissoring (bending)
F-Cl-F symmetric scissoring (bending)
F-Cl-F asymmetric scissoring (bending)
Cl-F symmetric stretching
Cl-F asymmetric stretching
Cl-F asymmetric stretching
Vibrational modes 1 and 2 are degenerate modes. However, since ClF3 molecule is not perfectly symmetric (It is not perfectly T-shaped), their vibrational frequencies are different.
There are three peaks seen in the IR spectrum. One broad peak at around 307 cm-1, one peak at around 735cm-1 and one very sharp peak at around 762cm-1.
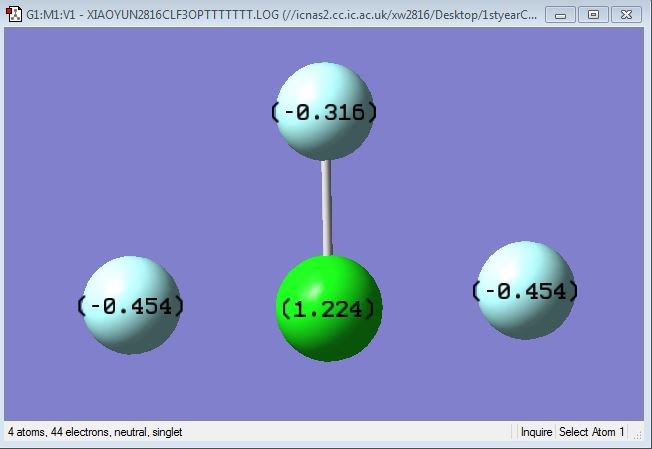
Charges on ClF3 molecule
Charge on Cl atom: 1.224 e-
Charge on F atom: -0.316 and -0.454 e-
Flourine atom is more electronegative than Chlorine atom. Thus F atoms are expected to have negative charges while Cl atom is expected to have a positive charge.
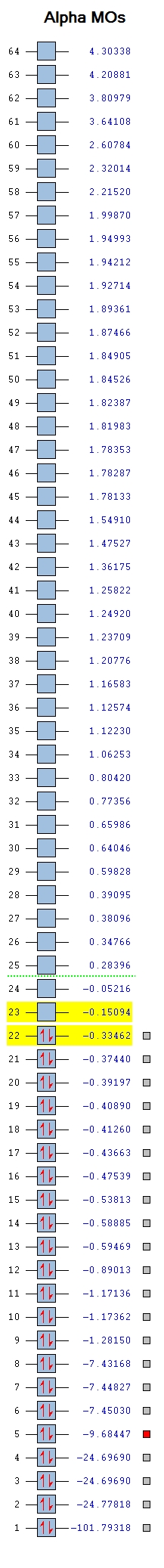
Molecular Orbitals of ClF3 molecule
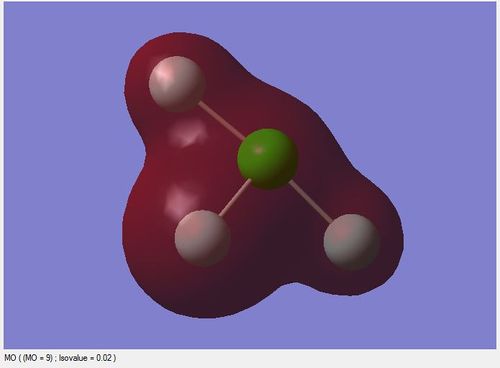
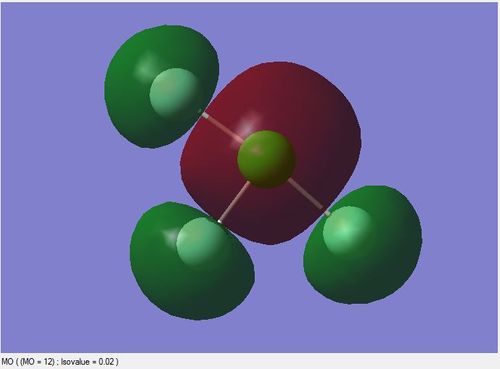
The image below is the Molecular Orbital(MO) table of ClF3 molecule
2s orbital of Cl
This molecular orbital is formed from the 2s orbital of Cl atom. It is very low in energy(-9.68447 au) due to the high effective nuclear charge of Cl.
It is a non-bonding molecular orbital filled with a pair of electrons.
It does not contribute in bonding as it is not a valence orbital and the energy difference between this orbital and 2s orbital of F is large.
2p orbital of Cl
This molecular orbital is formed by 2p orbital of Cl atom. It is low in energy(-7.43168 au) but higher in energy than the 2s orbital since it is less penetrating.
There are three degenerate molecular orbitals of the same type contributed by three degenerate 2p orbitals of the Cl atom.
It is a non-bonding molecular orbital filled with a pair of electrons.
It does not contribute in bonding as it is not a valence orbital and the energy difference between this orbital and 2p orbital of F is large.
bonding molecular orbital formed by 3s orbital of Cl and 2s orbitals of F
This is a molecular orbital formed by three 2s orbitals of F atoms and the 3s orbital of Cl atom. These atomic orbitals are all in phase with each other.
It is a bonding orbital filled with a pair of electrons. This is a sigma bonding orbital formed by head on overlaps of orbitals.
It is higher in energy (-1.28150 au) than the non-bonding orbitals.
anti-bonding molecular orbital formed by 3s orbital of Cl and 2s orbitals of F
This is a molecular orbital formed by three 2s orbitals of F atoms and the 3s orbital of Cl atom. The 3s orbital of Cl atom is out of phase with all three 2s orbitals of F atoms.
It is an anti-bonding molecular orbital filled with a pair of electrons. This is a sigma anti-bonding orbital formed by head on overlaps of orbitals.
It is higher in energy (-0.89013 au) than bonding molecular orbitals.
anti-bonding molecular orbital formed by 3p orbital of Cl and 2p orbitals of F
This is a molecular orbital formed by three 2p orbitals of F atoms and the 3p orbital of Cl atom. The 3p orbital of Cl atom is out of phase with all three 2p orbitals of F atoms.
It is an anti-bonding molecular orbital filled with a pair of electrons. This is a pi anti-bonding orbital formed by sideway overlaps of orbitals.
It is the highest in energy (-0.33462 au) among all occupied molecular orbitals. This is the highest occupied molecular orbital. (i.e.HOMO)
Reaction Energy of ClF3 molecule
Formation of ClF3 ː Cl2 + 3F2 = 2ClF3
E(ClF3) = -759.46531674 au
After optimising Cl2 and F2 with Gaussview, their energies are obtained.
E(Cl2)= -920.34987886 au
E(F2)= -199.49825218 au
ΔE=2×E(ClF3)-[E(Cl2)+3×E(F2)]= -759.46531674×2-(-920.34987886-3×199.49825218) = -0.08599808 au = -0.08599808×2625.5 kJ/mol = -225.79 kJ/mol (2d.p.)
The enthalpy change of formation of ClF3 is -225.79 kJ/mol. Since the reaction is exothermic, the product ClF3 is more stable than gaseous reactants.
The optimisation file of Cl2 is linked to here
The optimisation file of F2 is linked to here
References
- ↑ Haynes, W. M. (Ed.). (2014). CRC handbook of chemistry and physics. CRC press.
- ↑ Greenwood, N. N., & Earnshaw, A. (1997). Chemistry of the elements, Butterworth Heinemann. Oxford, ISBN, 80379419, 795.
- ↑ Cottrell, T. L. (1958). The strengths of chemical bonds. Academic Press.
- ↑ Modak, J. M. (2002). Haber process for ammonia synthesis. Resonance, 7(9), 69-77.