Rep:Mod: Sa13018
NH3 Molecule
test molecule (NH3) |
General
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES Predicted change in Energy=-5.986293D-10
Calculation Method RB3LYP Basis Set 6-31G(d,p) E(RB3LYP) -56.55776873 a.u. Dipole Moment 1.5008 Debye Point Group C3V
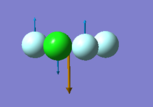
N-H bond distance = 1.02 a.u H-N-H bond angle = 106
Vibration analysis
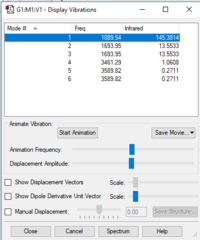
| wavenumber cm^-1 | 1090 | 1694 | 1694 | 3461 | 3590 | 3590 |
|---|---|---|---|---|---|---|
| symmetry | A1 | E | E | A1 | E | E |
| intensity(arbitary units) | 145 | 14 | 14 | 1 | 0 | 0 |
| Image |  |
 |
 |
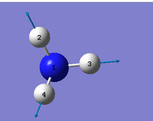 |
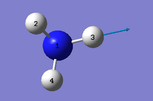 |
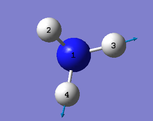
|
Number of modes= 6 Degenerate modes= modes at 1694, and 3590 cm^-1 bending vibrations= 1090,1694 stretching vibrations= 3461,3590 highly symmetric = 3461, and also 1090 (not as symmetric as 3461) umbrella mode= 1090 bands in gaseous ammonia= 3 The number of bands is 3, as there are 2 that don't result in a change in dipole moment, and so do not show an IR peak. There are then 2 degenerate peaks; they will show the same wavenumber, and so there are only 3 visibile bands in gaseous ammonia. For condensed it might be different, considering the effect of disorder.
Charge output
Charge on N = -1.125 Charge on H = 0.375 This seems like a likely charge distribution, considering the electronegativity of Nitrogen is much greater than Hydrogen, resulting in a greater electron density around the nitrogen, and an overall much greater charge (x3).
N2 molecule
General
test molecule (N2) |
Item Value Threshold Converged? Maximum Force 0.000145 0.000450 YES RMS Force 0.000145 0.000300 YES Maximum Displacement 0.000045 0.001800 YES RMS Displacement 0.000064 0.001200 YES Predicted change in Energy=-6.585141D-09
Calculation Method RB3LYP Basis Set 6-31G(d,p) E(RB3LYP) -109.52412867 a.u. Dipole Moment 0.0000 Debye Point Group D*H
Vibration analysis
N-N bond length= 1.11 a.u N-N bond angle= n/a. There are only 2 atoms, so no valid bond angle.
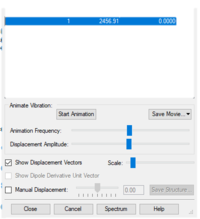
| wavenumber cm^-1 | 2457 |
|---|---|
| symmetry | SGG |
| intensity(arbitary units) | 0 |
| Image | 
|
Vibrational modes expected from 3N-5 = 1 Degenerate modes = n/a bending vibrations = n/a bond stretch vibrations = 2457 highly symmetric stretch = 2457 umbrella mode = n/a how many bands= zero; no change of dipole in the stretch
TM co-ordination
N2 is co-ordinated to an iridium complex, in ''bis((adamantan-1-ylmethyl)(diisopropyl)phosphine)-(dinitrogen)-hydrido-iridium''. The unique identifier for this is ANAZEV, and the article for the molecule can be found here: https://pubs.acs.org/doi/10.1021/ja1037808. The NN bond length in this molecule is 1.108 Å. This is almost identical to the bond length for the calculated computational length of 1.11. This implies that the computational calculation works and has found the correct stationary point for the optimised bond length. However, there is still a chance of some error; there is a possibility that the bond length could be shorter in reality than the computed distance, as the computed distance could be the result of a local minimum potential energy rather than the global minimum. If the bond length is shorter in reality, it would imply that the bond length of the co-ordinated NN bond is greater than computed. This would make sense as the electron density of the NN bond could be reduced by coordination, resulting in an overall longer bond. The link to the CCDC structure is this:https://www.ccdc.cam.ac.uk/structures/Search?Ccdcid=ANAZEV&DatabaseToSearch=Published
Charge output
There is no charge on the atoms, as the electronegativities are equal.
H2 molecule
General
test molecule (H2) |
Item Value Threshold Converged? Maximum Force 0.000066 0.000450 YES RMS Force 0.000066 0.000300 YES Maximum Displacement 0.000087 0.001800 YES RMS Displacement 0.000123 0.001200 YES Predicted change in Energy=-5.726834D-09
H-H bond length= 0.74 a.u H-H bond angle= n/a. There are only 2 molecules.
Calculation Method RB3LYP Basis Set 6-31G(d,p) E(RB3LYP) -1.17853935 a.u. Dipole Moment 0.0000 Debye Point Group D*H
Vibration analysis
| wavenumber cm^-1 | 4464 |
|---|---|
| symmetry | SGG |
| intensity(arbitary units) | 0 |
| Image | 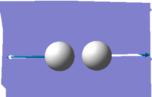
|
Vibrational modes expected from 3N-5 = 1 Degenerate modes = n/a bending vibrations = n/a bond stretch vibrations = 4464 highly symmetric stretch = 4464 umbrella mode = n/a how many bands= zero; no change of dipole in the stretch
Charge output

There is no charge on the atoms, as the electronegativities are equal.
The Haber Process
E(NH3)=-56.55776873 a.u 2*E(NH3)= -113.11553746 a.u E(N2)=-109.52413 a.u E(H2)=-1.17854 a.u 3*E(H2)= -3.53562 a.u ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= 0.05578746 a.u This is equal to -146.5 kJ/mol The process of converting N2 and 3H2 to 2NH3 is exothermic, and this implies that the more stable is the NH3 product over the reactants.
Project molecule (ClF3)
test molecule (ClF3) |
General
Item Value Threshold Converged? Maximum Force 0.000050 0.000450 YES RMS Force 0.000028 0.000300 YES Maximum Displacement 0.000204 0.001800 YES RMS Displacement 0.000134 0.001200 YES Predicted change in Energy==-1.250231D-08
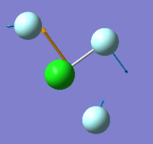
Cl-F(1) bond length= 1.65 a.u Cl-F(4) bond length= 1.73 a.u F4-Cl-F2 bond angle= 174 F4-Cl-F3 bond angle= 87
Calculation Method RB3LYP Basis Set 6-31G(d,p) E(RB3LYP) -759.46531688 a.u. Dipole Moment 0.8386 Debye Point Group C2V
Vibration analysis
| wavenumber cm^-1 | 305 | 309 | 401 | 541 | 736 | 752 | |
|---|---|---|---|---|---|---|---|
| symmetry | A1 | B1 | B2 | B2 | A1 | A1 | |
| intensity(arbitary units) | 14 | 18 | 1 | 541 | 736 | 752 | |
| Image |  |
 |
 |
 |
 |

|
Vibrational modes expected from 3N-6= 6 Degenerate modes = n/a bending vibrations = 305,309,401 bond stretch vibrations = 541,736,752 highly symmetric stretch = 541 umbrella mode = 309 how many bands= 5 There are 5 different vibrational modes that result in a change in dipole moment, so would cause 5 bands to appear in gaseous sample. The condensed state would not show this many, as there is more disorder to account for, and this would result in overlap between the closely lying bands.
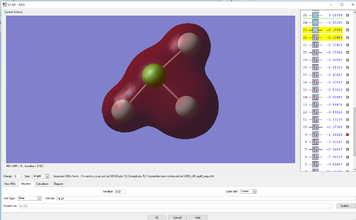
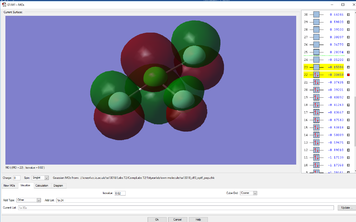
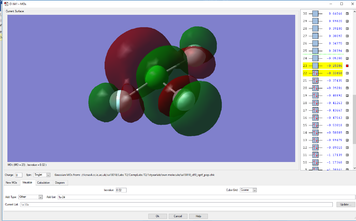
Molecular Orbital Analysis
 |
 |
||
 |
 |
||
 |
 |
Charge output
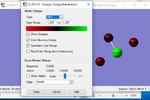
The F atoms are more electronegative than Cl, so have the negative charge. Cl has a charge of 1.225, and the individual F atoms have different charges based on position. The F3 middle atom is slightly less charged, with a charge of -0.315 as there is more repulsion of electrons from here due to increased electron density in that region. The outer F atoms have a charge of -0.465.
- ↑ This is based on works from this article by Henry Rzepa in the Winnower.https://thewinnower.com/papers/vsepr-theory-a-closer-look-at-chlorine-trifluoride-clf3
Marking
Note: All grades and comments are provisional and subject to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have received your grade from blackboard.
Wiki structure and presentation 0/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES, however you have written lots of text in the "pre" boxes, and you have not inserted line breaks leaving the reader to scroll sideways to read your work. You don't need to use the boxes the default formatting is much better.
Do you effectively use tables, figures and subheadings to communicate your work?
Lots of your tables are not formatted nicely and have extra rows/columns or no lines. Many of your figures are not cropped and one has been crudely coloured around in paint instead.
NH3 1/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES
N2 and H2 0.5/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES
Haber-Bosch reaction energy calculation 1/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 3.5/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
YES, good detailed explanations, well done. MO9 is made up of s type AOs not p type and the bonding is sigma bonding.
Independence 0/1
If you have finished everything else and have spare time in the lab you could: Check one of your results against the literature, or Do an extra calculation on another small molecule, or Do some deeper analysis on your results so far
No independent work.
